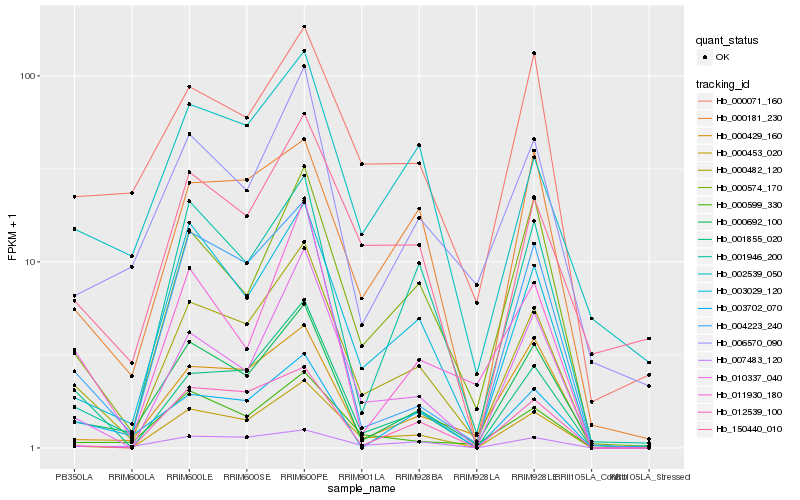
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000482_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 2 |
Hb_000453_020 |
0.1195830925 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000692_100 |
0.1216807299 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_003029_120 |
0.1250676537 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 5 |
Hb_001855_020 |
0.1253432236 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000574_170 |
0.1326998645 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 7 |
Hb_010337_040 |
0.1330500015 |
- |
- |
- |
| 8 |
Hb_007483_120 |
0.1421419872 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 9 |
Hb_000599_330 |
0.1595525026 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 10 |
Hb_000181_230 |
0.1622817022 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_150440_010 |
0.1632930008 |
- |
- |
- |
| 12 |
Hb_000071_160 |
0.1662839836 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 13 |
Hb_002539_050 |
0.1664444151 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 14 |
Hb_001946_200 |
0.1676875593 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_006570_090 |
0.1708192254 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 16 |
Hb_004223_240 |
0.1716460098 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 17 |
Hb_000429_160 |
0.1740807012 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 18 |
Hb_011930_180 |
0.1745717054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012539_100 |
0.1758979433 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 20 |
Hb_003702_070 |
0.1759179748 |
- |
- |
unnamed protein product [Vitis vinifera] |