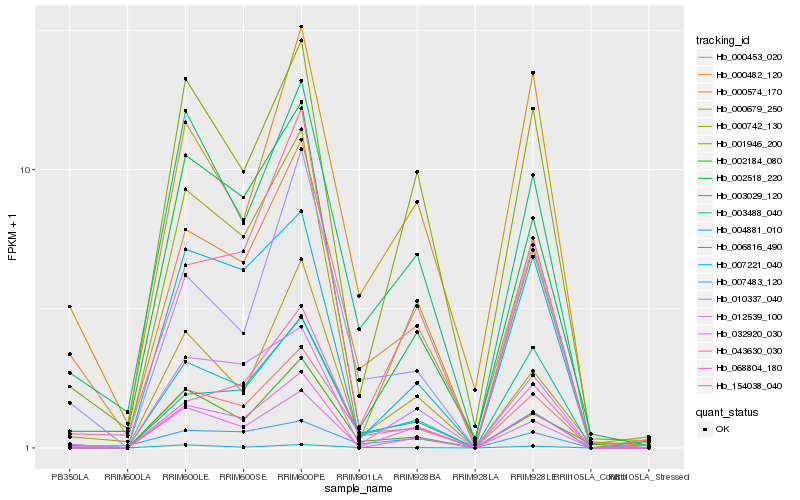
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_020 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000482_120 |
0.1195830925 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 3 |
Hb_003029_120 |
0.1220355587 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 4 |
Hb_010337_040 |
0.1250939939 |
- |
- |
- |
| 5 |
Hb_003488_040 |
0.1366763308 |
transcription factor |
TF Family: NF-YB |
nuclear transcription factor YB7 [Populus nigra x Populus x canadensis] |
| 6 |
Hb_002184_080 |
0.1404992902 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 7 |
Hb_012539_100 |
0.143561878 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 8 |
Hb_068804_180 |
0.1437823203 |
- |
- |
PREDICTED: TRAF-type zinc finger domain-containing protein 1 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_006816_490 |
0.1490522368 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 21 [Jatropha curcas] |
| 10 |
Hb_002518_220 |
0.1559175278 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001946_200 |
0.1575305289 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_000679_250 |
0.1578124155 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000742_130 |
0.1589342571 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 14 |
Hb_043630_030 |
0.1591542952 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 15 |
Hb_007221_040 |
0.159552442 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 16 |
Hb_000574_170 |
0.1612115895 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 17 |
Hb_007483_120 |
0.1632366605 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 18 |
Hb_154038_040 |
0.1637661189 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 19 |
Hb_004881_010 |
0.1656996321 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 20 |
Hb_032920_030 |
0.1663073244 |
- |
- |
signal transducer, putative [Ricinus communis] |