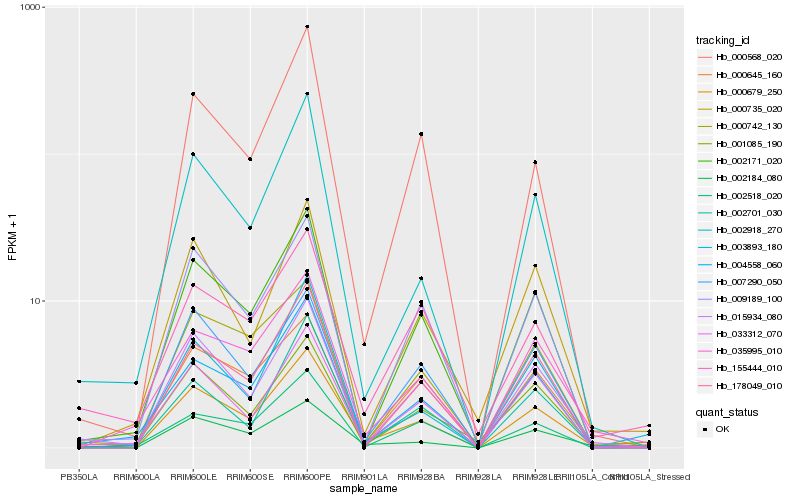
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 2 |
Hb_015934_080 |
0.1068873943 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 3 |
Hb_002171_020 |
0.1148720014 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_033312_070 |
0.1230626113 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 5 |
Hb_002184_080 |
0.1239372214 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 6 |
Hb_000645_160 |
0.1242108784 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 7 |
Hb_004558_060 |
0.1265179727 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 8 |
Hb_009189_100 |
0.1267672316 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 9 |
Hb_035995_010 |
0.1273218005 |
- |
- |
- |
| 10 |
Hb_178049_010 |
0.1275532678 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002918_270 |
0.1298947701 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 12 |
Hb_001085_190 |
0.1300861528 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 13 |
Hb_000735_020 |
0.1308671807 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_155444_010 |
0.1309703959 |
- |
- |
- |
| 15 |
Hb_002701_030 |
0.1320336546 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 16 |
Hb_000568_020 |
0.1332769626 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 17 |
Hb_003893_180 |
0.1335847139 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 18 |
Hb_007290_050 |
0.1338763279 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002518_020 |
0.1340481034 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 20 |
Hb_000742_130 |
0.1341400722 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |