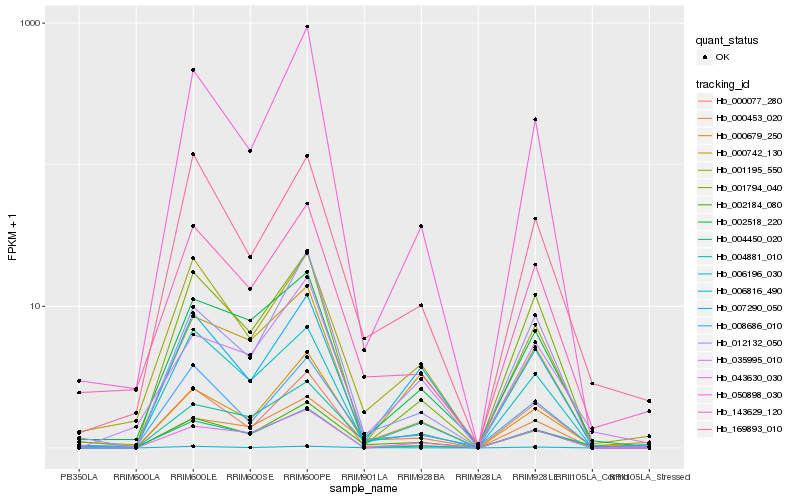
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002184_080 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_000679_250 |
0.1239372214 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002518_220 |
0.1371077399 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 4 |
Hb_169893_010 |
0.1389247592 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 5 |
Hb_006816_490 |
0.1394424194 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 21 [Jatropha curcas] |
| 6 |
Hb_000453_020 |
0.1404992902 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_035995_010 |
0.1444440142 |
- |
- |
- |
| 8 |
Hb_008686_010 |
0.1445908664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006196_030 |
0.1460642042 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 10 |
Hb_050898_030 |
0.1476677656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_143629_120 |
0.1488648524 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_043630_030 |
0.1518494559 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 13 |
Hb_004881_010 |
0.1519071176 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 14 |
Hb_001195_550 |
0.1533334697 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007290_050 |
0.1533584103 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004450_020 |
0.1534167942 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_012132_050 |
0.1549091988 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 18 |
Hb_000742_130 |
0.1551885284 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 19 |
Hb_001794_040 |
0.1554169122 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000077_280 |
0.1567582841 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |