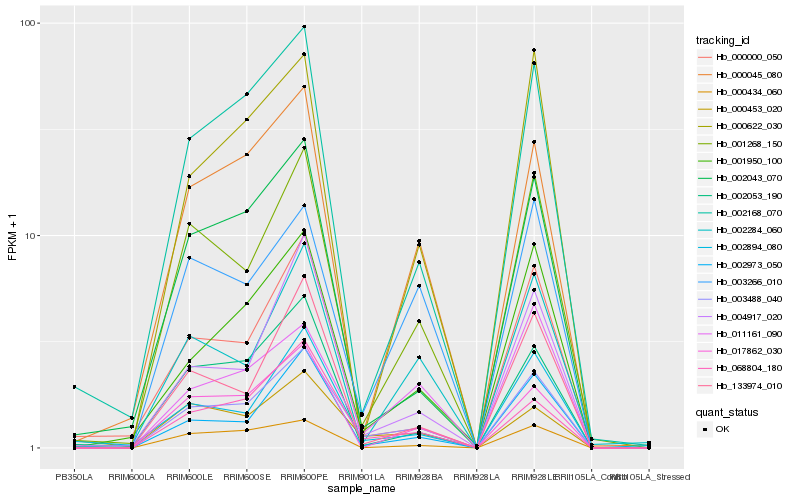
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003488_040 |
0.0 |
transcription factor |
TF Family: NF-YB |
nuclear transcription factor YB7 [Populus nigra x Populus x canadensis] |
| 2 |
Hb_001268_150 |
0.1190555296 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002973_050 |
0.1242523471 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 4 |
Hb_068804_180 |
0.1277189137 |
- |
- |
PREDICTED: TRAF-type zinc finger domain-containing protein 1 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001950_100 |
0.129810142 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 6 |
Hb_000453_020 |
0.1366763308 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002168_070 |
0.1380726801 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_004917_020 |
0.1411725396 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_133974_010 |
0.1415447365 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000045_080 |
0.1422894377 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_017862_030 |
0.1440932702 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 12 |
Hb_002894_080 |
0.1472489999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002284_060 |
0.1484104536 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_003266_010 |
0.1488595794 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 15 |
Hb_002043_070 |
0.1490558728 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 16 |
Hb_002053_190 |
0.1501342881 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011161_090 |
0.1515284182 |
- |
- |
- |
| 18 |
Hb_000000_050 |
0.15154479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000622_030 |
0.1516569708 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 20 |
Hb_000434_060 |
0.1522461579 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |