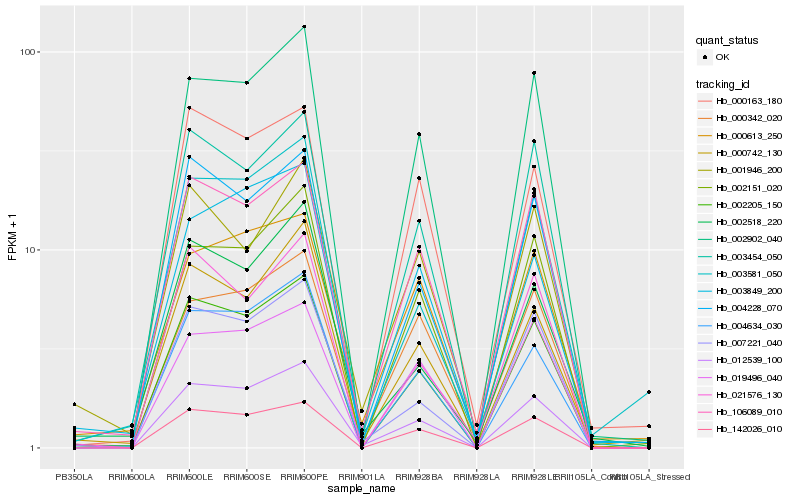
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012539_100 |
0.0 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 2 |
Hb_002205_150 |
0.0647486355 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 3 |
Hb_004634_030 |
0.0906657174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_142026_010 |
0.0930243538 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 5 |
Hb_106089_010 |
0.0936967271 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 6 |
Hb_002902_040 |
0.094786225 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 7 |
Hb_007221_040 |
0.099857786 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 8 |
Hb_000742_130 |
0.1015887048 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 9 |
Hb_003454_050 |
0.1020317178 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 10 |
Hb_002151_020 |
0.1021781662 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 11 |
Hb_004228_070 |
0.1087307406 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 12 |
Hb_002518_220 |
0.1129876894 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003849_200 |
0.1139189268 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_000342_020 |
0.1170538777 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 15 |
Hb_003581_050 |
0.1191662095 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 16 |
Hb_021576_130 |
0.1217464523 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000613_250 |
0.1229347484 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 18 |
Hb_000163_180 |
0.1252968881 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 19 |
Hb_019496_040 |
0.126159647 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 20 |
Hb_001946_200 |
0.1264138476 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |