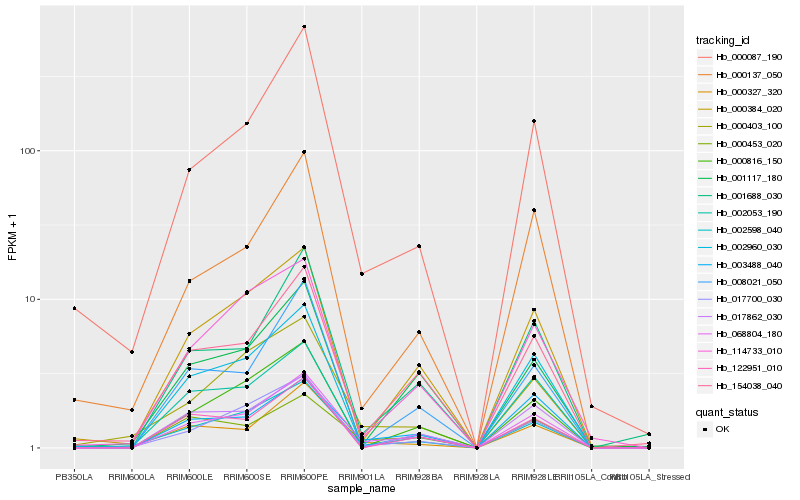
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_180 |
0.0 |
- |
- |
PREDICTED: TRAF-type zinc finger domain-containing protein 1 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_002598_040 |
0.1012235734 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 3 |
Hb_154038_040 |
0.113633957 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 4 |
Hb_017700_030 |
0.1186981565 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 5 |
Hb_001688_030 |
0.119754488 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 6 |
Hb_000816_150 |
0.1209138255 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 7 |
Hb_000384_020 |
0.1211887819 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 8 |
Hb_003488_040 |
0.1277189137 |
transcription factor |
TF Family: NF-YB |
nuclear transcription factor YB7 [Populus nigra x Populus x canadensis] |
| 9 |
Hb_002960_030 |
0.1301465381 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 10 |
Hb_008021_050 |
0.1306341157 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000087_190 |
0.1334761588 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 12 |
Hb_122951_010 |
0.1349376598 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 13 |
Hb_017862_030 |
0.1360297167 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 14 |
Hb_114733_010 |
0.1378890135 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001117_180 |
0.1405718394 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000327_320 |
0.1409717483 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g39360 [Jatropha curcas] |
| 17 |
Hb_000403_100 |
0.1414760667 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 18 |
Hb_000137_050 |
0.1419450895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002053_190 |
0.1420191075 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000453_020 |
0.1437823203 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |