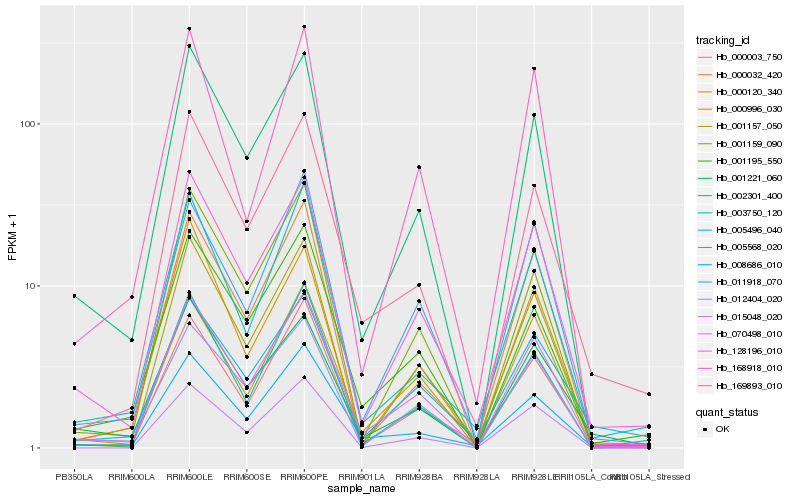
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070498_010 |
0.0 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 2 |
Hb_128196_010 |
0.0712038394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002301_400 |
0.074651697 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_000996_030 |
0.0805168994 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012404_020 |
0.0808723966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011918_070 |
0.0845543113 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 7 |
Hb_000003_750 |
0.0869016205 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 8 |
Hb_001195_550 |
0.08990064 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008686_010 |
0.0913322999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001157_050 |
0.0941001267 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 11 |
Hb_169893_010 |
0.0941187946 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 12 |
Hb_015048_020 |
0.0993170096 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 13 |
Hb_003750_120 |
0.1003900531 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 14 |
Hb_001221_060 |
0.1048838827 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 15 |
Hb_001159_090 |
0.1059719068 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 16 |
Hb_168918_010 |
0.106501138 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_005568_020 |
0.1082059858 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000032_420 |
0.1087923521 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 19 |
Hb_005496_040 |
0.110112915 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 20 |
Hb_000120_340 |
0.1124191083 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |