| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
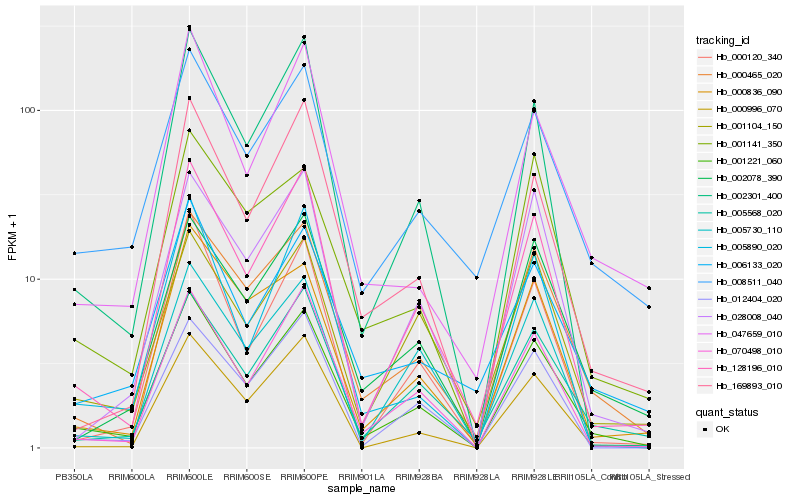
Hb_001221_060 |
0.0 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 2 |
Hb_128196_010 |
0.0884532644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002301_400 |
0.1005515968 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_000465_020 |
0.1043998855 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 5 |
Hb_070498_010 |
0.1048838827 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 6 |
Hb_005568_020 |
0.1074946586 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008511_040 |
0.1167842458 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_002078_390 |
0.1182548426 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 9 |
Hb_000120_340 |
0.1183337991 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_000836_090 |
0.1207086144 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 11 |
Hb_169893_010 |
0.1223001701 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 12 |
Hb_012404_020 |
0.1228791749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_047659_010 |
0.1249606584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006133_020 |
0.1265026607 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 15 |
Hb_005890_020 |
0.1279016801 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_028008_040 |
0.1286918631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001104_150 |
0.1306787671 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 18 |
Hb_000996_070 |
0.1314944279 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001141_350 |
0.1318227306 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 20 |
Hb_005730_110 |
0.1324325559 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |