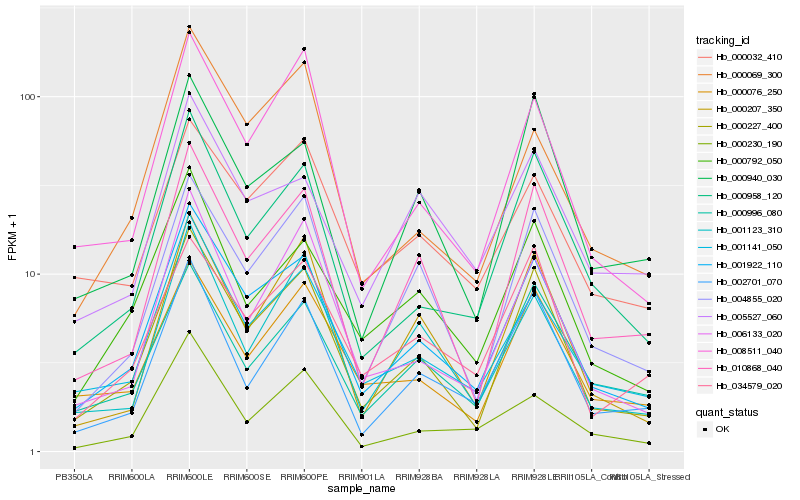
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001922_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000227_400 |
0.0660183394 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 3 |
Hb_001141_050 |
0.0823777101 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000958_120 |
0.0959895475 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 5 |
Hb_000792_050 |
0.107712771 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_010868_040 |
0.1077444947 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 7 |
Hb_008511_040 |
0.1128039951 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_004855_020 |
0.117060978 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 9 |
Hb_006133_020 |
0.1171205353 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 10 |
Hb_000069_300 |
0.12209586 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000032_410 |
0.1233231814 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 12 |
Hb_000076_250 |
0.124339266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000207_350 |
0.1266186367 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 14 |
Hb_002701_070 |
0.1277267207 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 15 |
Hb_000940_030 |
0.1305599197 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005527_060 |
0.1316467977 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 17 |
Hb_034579_020 |
0.1320629093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000996_080 |
0.1326342978 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 19 |
Hb_000230_190 |
0.132728966 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001123_310 |
0.1329028421 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |