| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
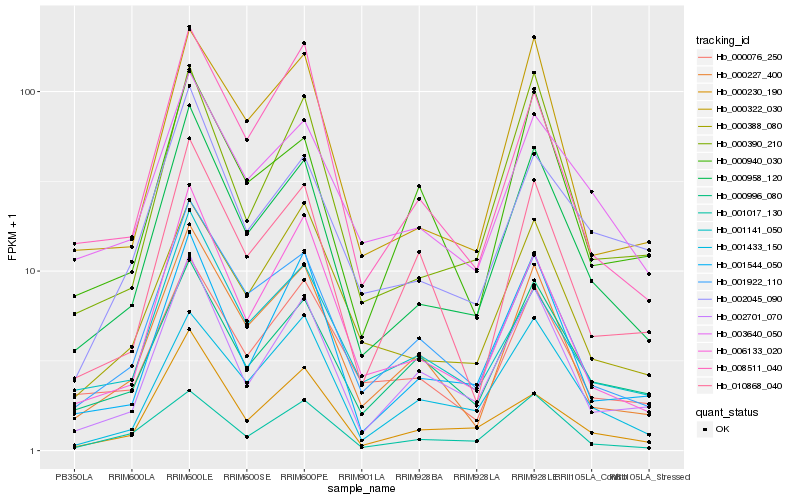
Hb_000958_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 2 |
Hb_001922_110 |
0.0959895475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002701_070 |
0.1024900976 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 4 |
Hb_000227_400 |
0.1026746278 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 5 |
Hb_000322_030 |
0.1095827506 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 6 |
Hb_000390_210 |
0.1099579131 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_001544_050 |
0.114392763 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 8 |
Hb_006133_020 |
0.1194793767 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 9 |
Hb_001141_050 |
0.1204868631 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000996_080 |
0.1228317712 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 11 |
Hb_003640_050 |
0.1251439889 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_002045_090 |
0.1262276642 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 13 |
Hb_000230_190 |
0.1268885714 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000076_250 |
0.1293453018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010868_040 |
0.1298954427 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 16 |
Hb_000388_080 |
0.1305317507 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 17 |
Hb_008511_040 |
0.1325181531 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_000940_030 |
0.1334820203 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001017_130 |
0.1343615205 |
- |
- |
hypothetical protein POPTR_0010s06070g [Populus trichocarpa] |
| 20 |
Hb_001433_150 |
0.1348903688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |