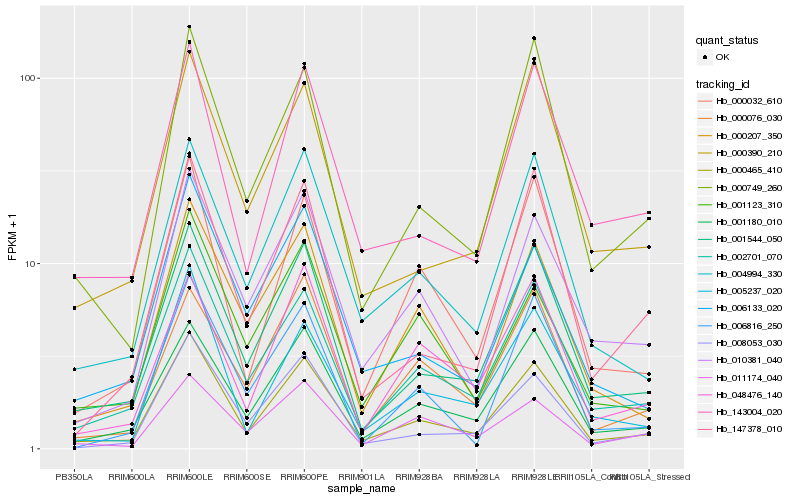
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_410 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002701_070 |
0.070043217 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 3 |
Hb_001544_050 |
0.0747582422 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_001180_010 |
0.0858226522 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 5 |
Hb_000032_610 |
0.0947312012 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_004994_330 |
0.0978707284 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000749_260 |
0.0995017486 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005237_020 |
0.1030873388 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_143004_020 |
0.1038627999 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 10 |
Hb_008053_030 |
0.1062700046 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 11 |
Hb_010381_040 |
0.1132506548 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 12 |
Hb_147378_010 |
0.1157987891 |
- |
- |
- |
| 13 |
Hb_000390_210 |
0.1174076096 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_000207_350 |
0.1186367693 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 15 |
Hb_006133_020 |
0.1248655288 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 16 |
Hb_048476_140 |
0.1268398692 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 17 |
Hb_001123_310 |
0.1308760466 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 18 |
Hb_000076_030 |
0.1323027884 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011174_040 |
0.1323408946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006816_250 |
0.1347970465 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |