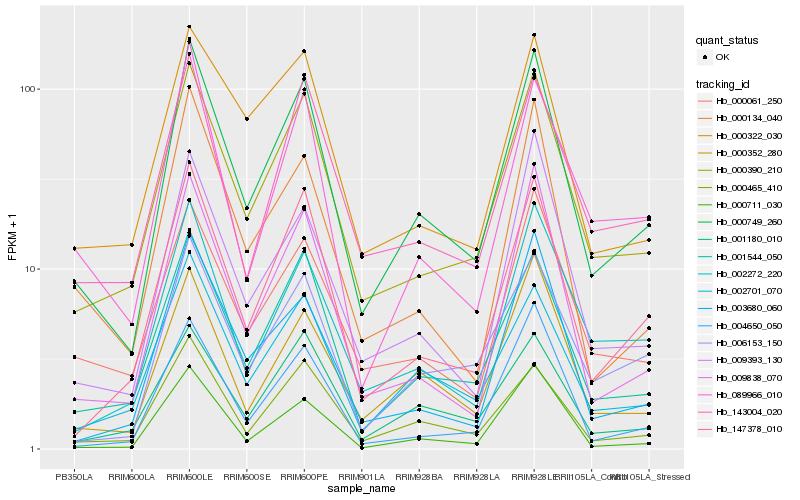
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_260 |
0.0 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000390_210 |
0.0812610333 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_001544_050 |
0.0900679743 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_147378_010 |
0.0948884972 |
- |
- |
- |
| 5 |
Hb_000465_410 |
0.0995017486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002701_070 |
0.1056399418 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 7 |
Hb_000352_280 |
0.1089793485 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 8 |
Hb_009838_070 |
0.1094948816 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006153_150 |
0.115715053 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 10 |
Hb_143004_020 |
0.1172439598 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 11 |
Hb_001180_010 |
0.1173886351 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 12 |
Hb_000711_030 |
0.1185768688 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 13 |
Hb_000061_250 |
0.1197950292 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 14 |
Hb_089966_010 |
0.1210433097 |
- |
- |
- |
| 15 |
Hb_000134_040 |
0.1211506308 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002272_220 |
0.1212241559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009393_130 |
0.1216243938 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 18 |
Hb_004650_050 |
0.1230424064 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000322_030 |
0.124817649 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 20 |
Hb_003680_060 |
0.1263788959 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |