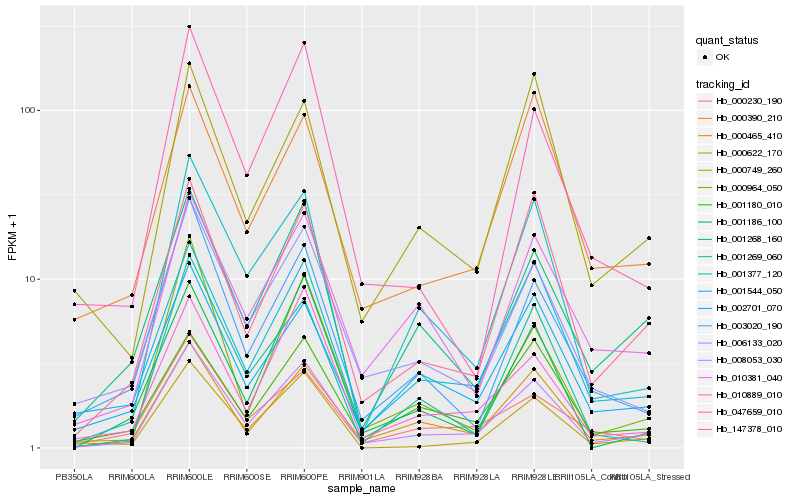
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008053_030 |
0.0 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 2 |
Hb_000465_410 |
0.1062700046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001544_050 |
0.1118213882 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_001377_120 |
0.1126470152 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 5 |
Hb_147378_010 |
0.1129547294 |
- |
- |
- |
| 6 |
Hb_006133_020 |
0.1165165198 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 7 |
Hb_001268_160 |
0.1257637858 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 8 |
Hb_000622_170 |
0.1284154442 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 9 |
Hb_002701_070 |
0.1302847654 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 10 |
Hb_010889_010 |
0.131802744 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 11 |
Hb_001269_060 |
0.1346664325 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010381_040 |
0.1359159606 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_001180_010 |
0.1376509925 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 14 |
Hb_003020_190 |
0.1379365555 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 15 |
Hb_000230_190 |
0.1383392996 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000964_050 |
0.1421655556 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 17 |
Hb_001186_100 |
0.1426495329 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 18 |
Hb_047659_010 |
0.146019732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000390_210 |
0.1460459735 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_000749_260 |
0.1461396793 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |