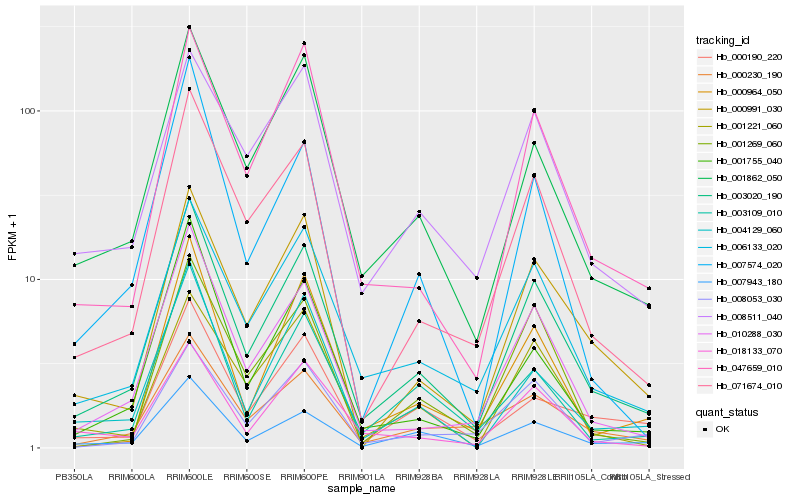
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_190 |
0.0 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 2 |
Hb_010288_030 |
0.0933574858 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_071674_010 |
0.0939729931 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 4 |
Hb_006133_020 |
0.0953446279 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 5 |
Hb_001862_050 |
0.1029439768 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 6 |
Hb_000991_030 |
0.1116042345 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 7 |
Hb_007943_180 |
0.1143097888 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 8 |
Hb_047659_010 |
0.1156257591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000230_190 |
0.1216416448 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004129_060 |
0.1224199857 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 11 |
Hb_018133_070 |
0.1296211559 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 12 |
Hb_008511_040 |
0.1298700645 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_001755_040 |
0.1318447239 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_060 |
0.1329608422 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 15 |
Hb_000964_050 |
0.133387635 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 16 |
Hb_001269_060 |
0.1341302178 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003109_010 |
0.1342497277 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 18 |
Hb_008053_030 |
0.1379365555 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 19 |
Hb_000190_220 |
0.138245561 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 20 |
Hb_007574_020 |
0.138597796 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |