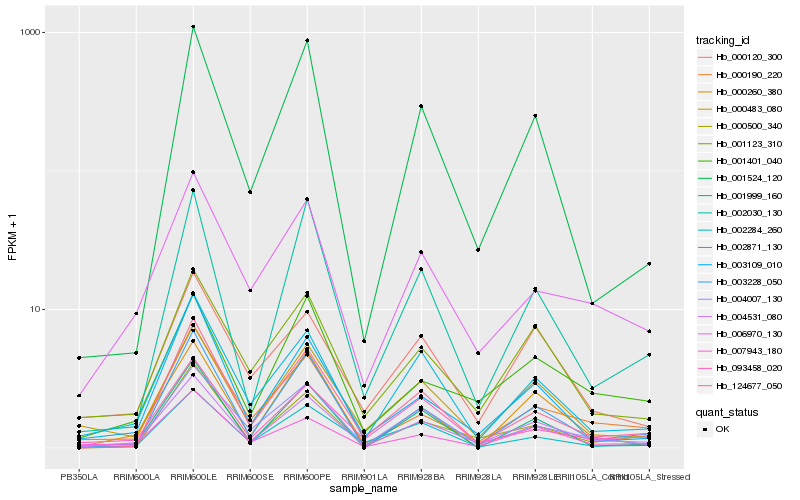
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_080 |
0.0 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 2 |
Hb_002871_130 |
0.1036993599 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 3 |
Hb_007943_180 |
0.1131533443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 4 |
Hb_000500_340 |
0.115242434 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 5 |
Hb_004007_130 |
0.1169977013 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 6 |
Hb_001123_310 |
0.1205515114 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 7 |
Hb_000483_080 |
0.1219707087 |
- |
- |
hypothetical protein CISIN_1g000833mg [Citrus sinensis] |
| 8 |
Hb_003109_010 |
0.1277304681 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 9 |
Hb_000190_220 |
0.1283448214 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 10 |
Hb_003228_050 |
0.1288748434 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 11 |
Hb_006970_130 |
0.1293395009 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 12 |
Hb_124677_050 |
0.1343856989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001524_120 |
0.1353987018 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_093458_020 |
0.1357454704 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 15 |
Hb_002284_260 |
0.1388751186 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 16 |
Hb_000120_300 |
0.1392904016 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 17 |
Hb_001401_040 |
0.1393809222 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 18 |
Hb_002030_130 |
0.1394293194 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 19 |
Hb_000260_380 |
0.1397300496 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 20 |
Hb_001999_160 |
0.1410975291 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |