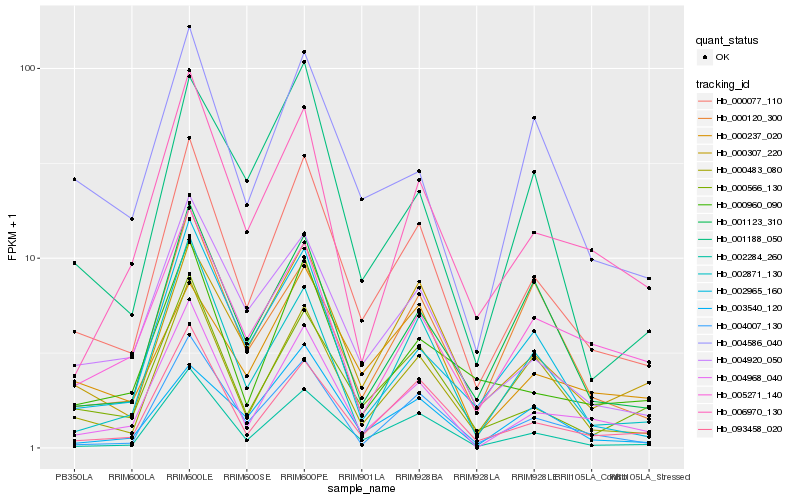
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 2 |
Hb_000483_080 |
0.1057376142 |
- |
- |
hypothetical protein CISIN_1g000833mg [Citrus sinensis] |
| 3 |
Hb_000307_220 |
0.1063632235 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 4 |
Hb_000566_130 |
0.1087499017 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 5 |
Hb_093458_020 |
0.1167974754 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 6 |
Hb_004920_050 |
0.1168060644 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 7 |
Hb_002284_260 |
0.1170583156 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 8 |
Hb_004968_040 |
0.1218275983 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 9 |
Hb_004007_130 |
0.132497587 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 10 |
Hb_000960_090 |
0.1412457422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001123_310 |
0.1417437599 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 12 |
Hb_006970_130 |
0.1433801381 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 13 |
Hb_003540_120 |
0.1441178847 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_002871_130 |
0.144913363 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 15 |
Hb_004586_040 |
0.145069036 |
transcription factor |
TF Family: MIKC |
K-box region and MADS-box transcription factor family protein [Theobroma cacao] |
| 16 |
Hb_000237_020 |
0.1475201538 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 17 |
Hb_002965_160 |
0.1479942548 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 18 |
Hb_001188_050 |
0.1528390047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000120_300 |
0.155878851 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 20 |
Hb_005271_140 |
0.1566275334 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |