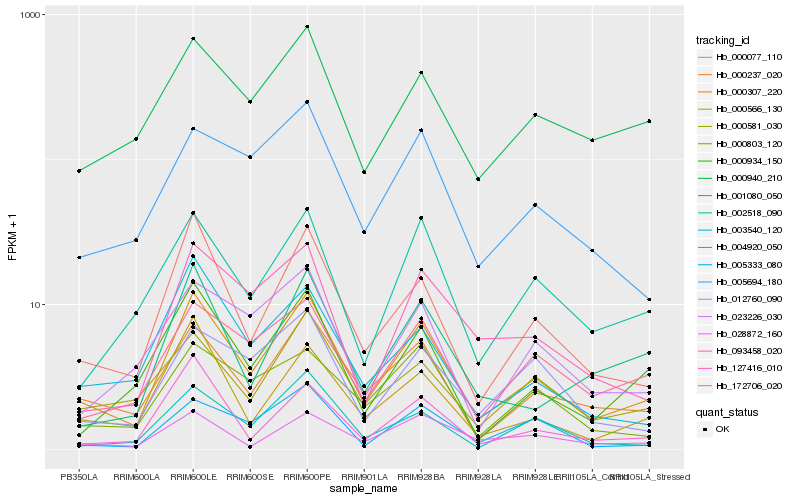
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_220 |
0.0 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 2 |
Hb_000077_110 |
0.1063632235 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 3 |
Hb_000237_020 |
0.1142700308 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 4 |
Hb_000566_130 |
0.1370982985 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 5 |
Hb_172706_020 |
0.1420920841 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 6 |
Hb_000581_030 |
0.1453506782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002518_090 |
0.1477222899 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 8 |
Hb_000934_150 |
0.1500688741 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 9 |
Hb_000940_210 |
0.1531682287 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 10 |
Hb_003540_120 |
0.1548079639 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_004920_050 |
0.154905514 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 12 |
Hb_005694_180 |
0.1550947241 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 13 |
Hb_093458_020 |
0.1567468599 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 14 |
Hb_127416_010 |
0.1572782319 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_012760_090 |
0.1594078599 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 16 |
Hb_005333_080 |
0.1594885798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023226_030 |
0.1603896134 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 18 |
Hb_028872_160 |
0.1614430571 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 19 |
Hb_000803_120 |
0.1618378297 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001080_050 |
0.1624119563 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |