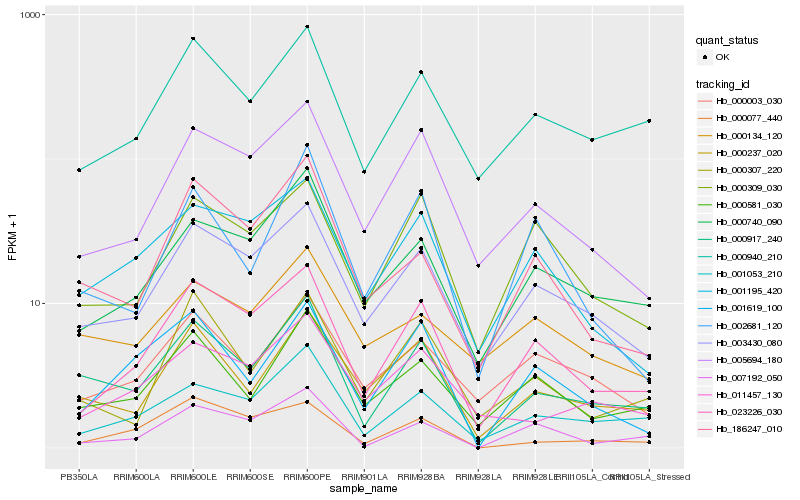
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_240 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 2 |
Hb_000237_020 |
0.1346228587 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 3 |
Hb_003430_080 |
0.1415222613 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_001053_210 |
0.1465103838 |
- |
- |
- |
| 5 |
Hb_000581_030 |
0.1527534163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000940_210 |
0.1578359893 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 7 |
Hb_023226_030 |
0.1589212313 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 8 |
Hb_000740_090 |
0.161892469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_186247_010 |
0.1621598946 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 10 |
Hb_005694_180 |
0.1696771637 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 11 |
Hb_000307_220 |
0.1721908267 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 12 |
Hb_001195_420 |
0.1730988492 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 13 |
Hb_011457_130 |
0.1781323933 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 14 |
Hb_007192_050 |
0.178790486 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 15 |
Hb_001619_100 |
0.1811025858 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 16 |
Hb_000003_030 |
0.1827098985 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000077_440 |
0.1848811999 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 18 |
Hb_002681_120 |
0.1861557477 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 19 |
Hb_000309_030 |
0.1863942082 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000134_120 |
0.1864587773 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |