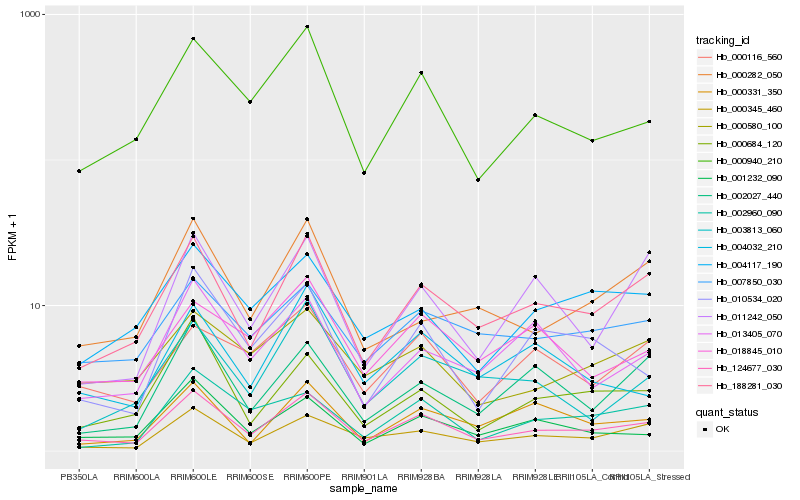
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124677_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 2 |
Hb_188281_030 |
0.0892052822 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 3 |
Hb_000940_210 |
0.1111048491 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 4 |
Hb_000345_460 |
0.1230424546 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 5 |
Hb_001232_090 |
0.1310014679 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 6 |
Hb_010534_020 |
0.1399355063 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000684_120 |
0.1419660492 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007850_030 |
0.1436341145 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 9 |
Hb_004117_190 |
0.1489809719 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 10 |
Hb_000580_100 |
0.1490314171 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000116_560 |
0.1492858464 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 12 |
Hb_013405_070 |
0.1496321822 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 13 |
Hb_004032_210 |
0.1501211869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002960_090 |
0.1511394594 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_002027_440 |
0.15119616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011242_050 |
0.1514915811 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 17 |
Hb_000331_350 |
0.1515854133 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 18 |
Hb_000282_050 |
0.1532732663 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 19 |
Hb_018845_010 |
0.1554027703 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 20 |
Hb_003813_060 |
0.1563534767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |