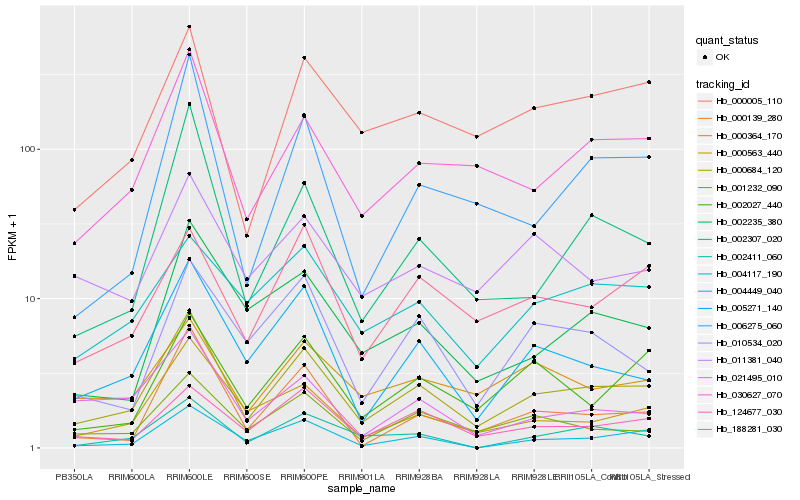
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 2 |
Hb_030627_070 |
0.1233938327 |
- |
- |
histone H4 [Zea mays] |
| 3 |
Hb_000563_440 |
0.124074519 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005271_140 |
0.131932665 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 5 |
Hb_001232_090 |
0.1352548131 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 6 |
Hb_124677_030 |
0.1419660492 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 7 |
Hb_004449_040 |
0.1435707296 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_000005_110 |
0.1455211144 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 9 |
Hb_002411_060 |
0.1503020184 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 10 |
Hb_002235_380 |
0.1551670646 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 11 |
Hb_000139_280 |
0.1559548936 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_188281_030 |
0.1577460509 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 13 |
Hb_000364_170 |
0.1581386574 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 14 |
Hb_002307_020 |
0.1581672013 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 15 |
Hb_006275_060 |
0.1584756216 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 16 |
Hb_021495_010 |
0.1616781027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002027_440 |
0.1628986082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004117_190 |
0.1629924107 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 19 |
Hb_011381_040 |
0.1639448261 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010534_020 |
0.1653707116 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |