| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
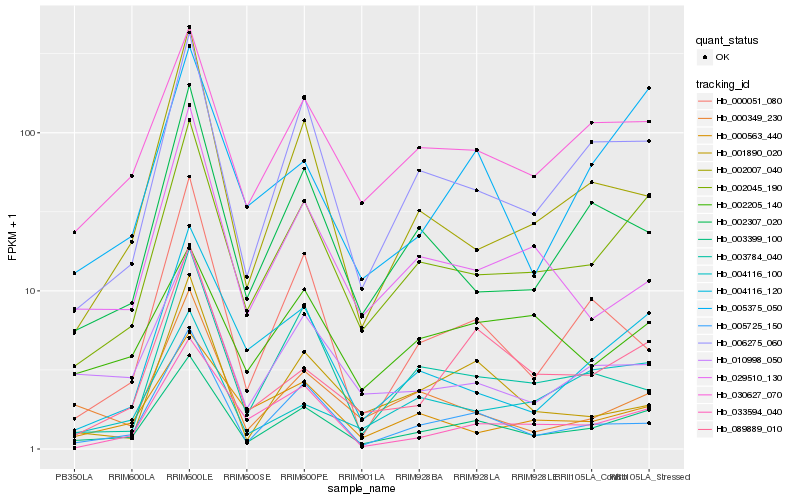
Hb_003399_100 |
0.0 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 2 |
Hb_002045_190 |
0.0997896767 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 3 |
Hb_030627_070 |
0.1202315565 |
- |
- |
histone H4 [Zea mays] |
| 4 |
Hb_006275_060 |
0.1279807209 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 5 |
Hb_005725_150 |
0.1370699169 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010998_050 |
0.1514734672 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 7 |
Hb_089889_010 |
0.1531070077 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_005375_050 |
0.1676487853 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 9 |
Hb_000349_230 |
0.1680018852 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 10 |
Hb_033594_040 |
0.170634929 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002307_020 |
0.171323067 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 12 |
Hb_004116_120 |
0.1745920362 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 13 |
Hb_000563_440 |
0.181621238 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003784_040 |
0.1834328519 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001890_020 |
0.1849768673 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 16 |
Hb_029510_130 |
0.1857648014 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 17 |
Hb_002205_140 |
0.1878856475 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 18 |
Hb_000051_080 |
0.1880774655 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 19 |
Hb_002007_040 |
0.188964386 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_004116_100 |
0.1892285886 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |