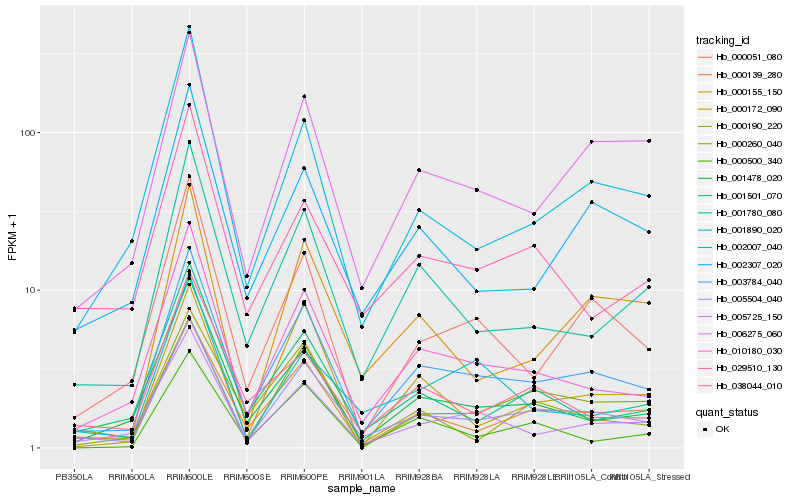
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003784_040 |
0.0 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 2 |
Hb_005504_040 |
0.0687471852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010180_030 |
0.0897189902 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 4 |
Hb_000139_280 |
0.1034080546 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005725_150 |
0.1078658041 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001478_020 |
0.1096376444 |
desease resistance |
Gene Name: AAA |
thyroid hormone receptor interactor, putative [Ricinus communis] |
| 7 |
Hb_001780_080 |
0.1100268943 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 8 |
Hb_000051_080 |
0.1128207349 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 9 |
Hb_006275_060 |
0.1232657796 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 10 |
Hb_002307_020 |
0.1314789288 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 11 |
Hb_002007_040 |
0.1336320567 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_001501_070 |
0.1408393887 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 13 |
Hb_038044_010 |
0.1458100431 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 14 |
Hb_000500_340 |
0.1500311427 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 15 |
Hb_000260_040 |
0.1526677229 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 16 |
Hb_029510_130 |
0.1547517493 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 17 |
Hb_000155_150 |
0.1551802296 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 18 |
Hb_000190_220 |
0.1598996243 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 19 |
Hb_001890_020 |
0.1636244919 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 20 |
Hb_000172_090 |
0.1665289866 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |