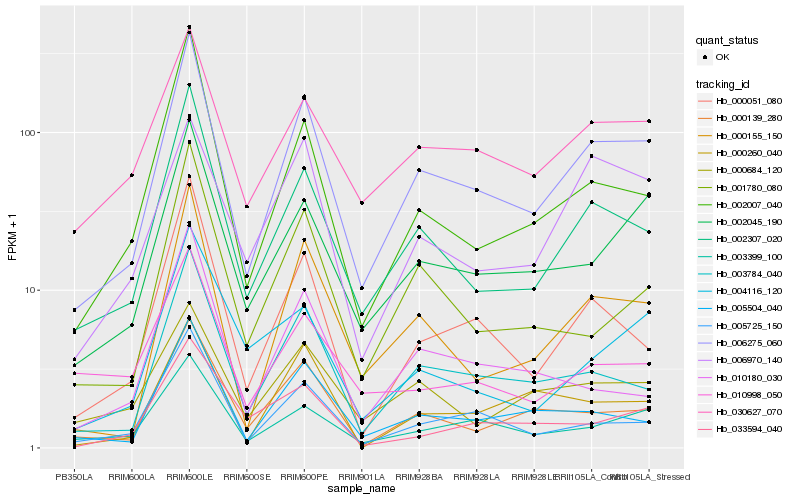
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006275_060 |
0.0 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 2 |
Hb_002307_020 |
0.1010330547 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 3 |
Hb_000155_150 |
0.1190852289 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 4 |
Hb_002045_190 |
0.1209985517 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 5 |
Hb_003784_040 |
0.1232657796 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 6 |
Hb_003399_100 |
0.1279807209 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 7 |
Hb_005725_150 |
0.1344138216 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005504_040 |
0.1356546481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000139_280 |
0.1370272637 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_030627_070 |
0.1387858809 |
- |
- |
histone H4 [Zea mays] |
| 11 |
Hb_000051_080 |
0.1394303572 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 12 |
Hb_002007_040 |
0.1435334029 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001780_080 |
0.1552609288 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 14 |
Hb_000684_120 |
0.1584756216 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000260_040 |
0.1648579587 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 16 |
Hb_004116_120 |
0.167055283 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 17 |
Hb_006970_140 |
0.1700052926 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 18 |
Hb_033594_040 |
0.1741745981 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_010180_030 |
0.1812787312 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 20 |
Hb_010998_050 |
0.1822939696 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |