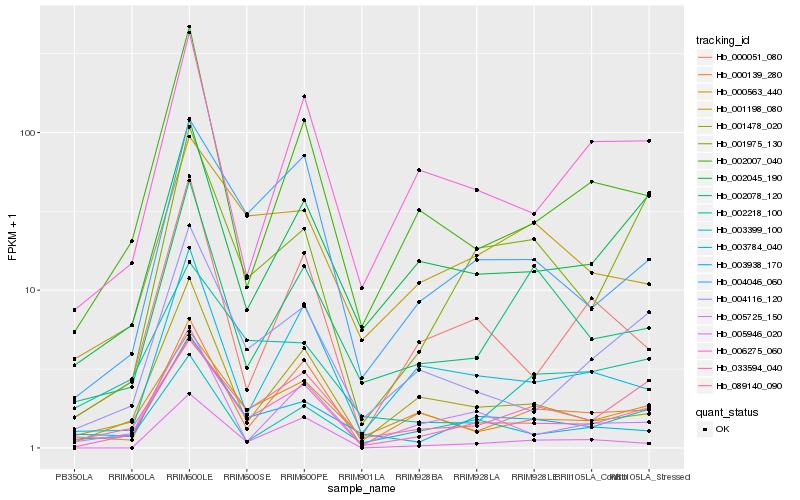
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_040 |
0.0 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_004046_060 |
0.1390459199 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 3 |
Hb_004116_120 |
0.142293471 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 4 |
Hb_089140_090 |
0.1424182051 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_002045_190 |
0.1544466337 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 6 |
Hb_000563_440 |
0.1554749702 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000139_280 |
0.1622511378 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001975_130 |
0.1701865381 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 9 |
Hb_003399_100 |
0.170634929 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 10 |
Hb_006275_060 |
0.1741745981 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 11 |
Hb_001478_020 |
0.1742745238 |
desease resistance |
Gene Name: AAA |
thyroid hormone receptor interactor, putative [Ricinus communis] |
| 12 |
Hb_005725_150 |
0.178982734 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003784_040 |
0.1812131765 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 14 |
Hb_002007_040 |
0.1840746342 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_003938_170 |
0.1858923925 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 16 |
Hb_001198_080 |
0.1901189852 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000051_080 |
0.1919695157 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 18 |
Hb_002218_100 |
0.1951097024 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 19 |
Hb_002078_120 |
0.1954752145 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 20 |
Hb_005946_020 |
0.1959241718 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |