| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
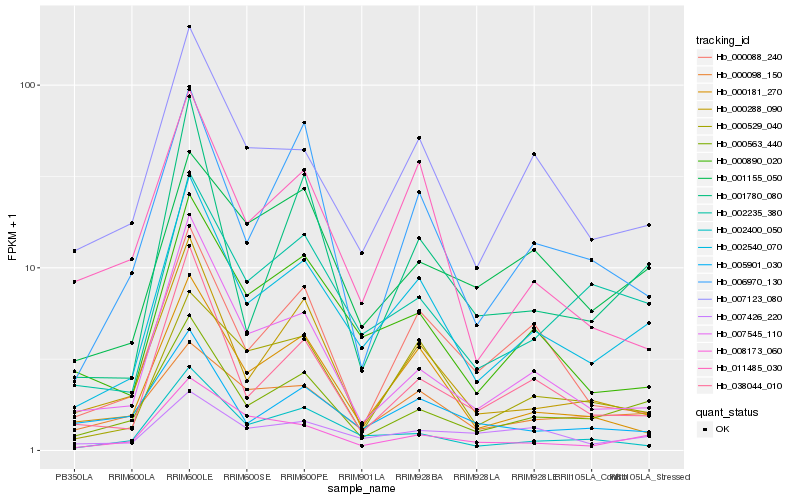
| 1 |
Hb_002540_070 |
0.0 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 2 |
Hb_007123_080 |
0.1226058393 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 3 |
Hb_000563_440 |
0.1233955123 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000890_020 |
0.1328677815 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 5 |
Hb_002235_380 |
0.136927572 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 6 |
Hb_000181_270 |
0.1391120955 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_002400_050 |
0.1481033682 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000529_040 |
0.1509756451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 9 |
Hb_038044_010 |
0.1572256094 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 10 |
Hb_007545_110 |
0.1586656484 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000288_090 |
0.1586918048 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_008173_060 |
0.15909068 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 13 |
Hb_001155_050 |
0.1654106974 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 14 |
Hb_000098_150 |
0.1671479413 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 15 |
Hb_007426_220 |
0.1697589503 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005901_030 |
0.1699009613 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011485_030 |
0.1700089085 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 18 |
Hb_001780_080 |
0.1703366525 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 19 |
Hb_006970_130 |
0.1706842264 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 20 |
Hb_000088_240 |
0.1709867293 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |