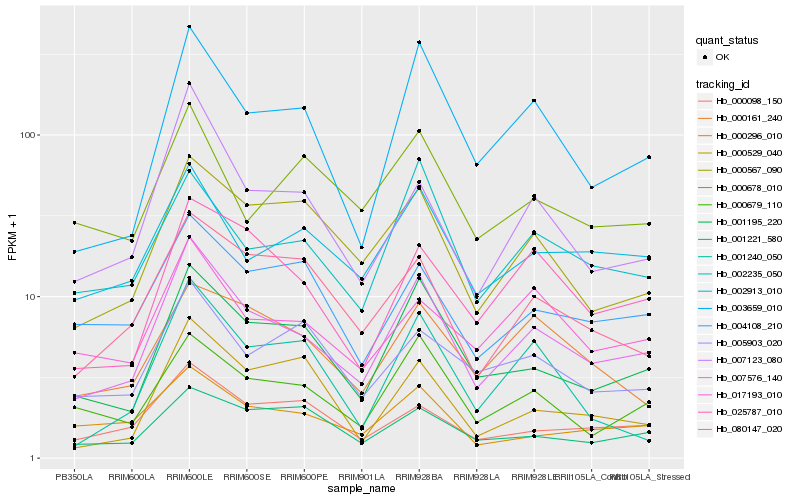
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_140 |
0.0 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 2 |
Hb_001195_220 |
0.1134378859 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_003659_010 |
0.1173459716 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 4 |
Hb_007123_080 |
0.1359542307 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 5 |
Hb_000679_110 |
0.136180747 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 6 |
Hb_025787_010 |
0.1365835007 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 7 |
Hb_000567_090 |
0.1379073813 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002913_010 |
0.1383128444 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002235_050 |
0.1416788671 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_080147_020 |
0.1450432607 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 11 |
Hb_017193_010 |
0.147860209 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000098_150 |
0.1500232544 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 13 |
Hb_004108_210 |
0.1502966964 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 14 |
Hb_000296_010 |
0.153570318 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 15 |
Hb_005903_020 |
0.159602416 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 16 |
Hb_001221_580 |
0.1602116531 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 17 |
Hb_000161_240 |
0.1621247854 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000529_040 |
0.1653825456 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 19 |
Hb_000678_010 |
0.165972338 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001240_050 |
0.1710814009 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |