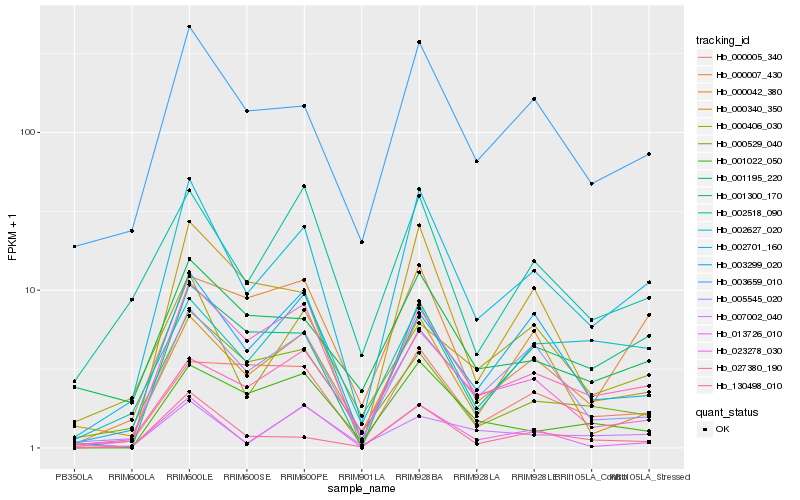
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_160 |
0.0 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 2 |
Hb_003659_010 |
0.1372485269 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 3 |
Hb_005545_020 |
0.1604106534 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 4 |
Hb_130498_010 |
0.1719607873 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 5 |
Hb_001300_170 |
0.1730390805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_013726_010 |
0.1747643482 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_003299_020 |
0.1796329474 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 8 |
Hb_023278_030 |
0.1826996152 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002518_090 |
0.1898785746 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 10 |
Hb_001195_220 |
0.1907657293 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_027380_190 |
0.1931940795 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000340_350 |
0.19355486 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001022_050 |
0.1946634525 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 14 |
Hb_000529_040 |
0.1970274618 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 15 |
Hb_000005_340 |
0.198326726 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007002_040 |
0.1995657387 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 17 |
Hb_000007_430 |
0.1996489385 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 18 |
Hb_000406_030 |
0.199760879 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 19 |
Hb_000042_380 |
0.2006002949 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 20 |
Hb_002627_020 |
0.2009260663 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |