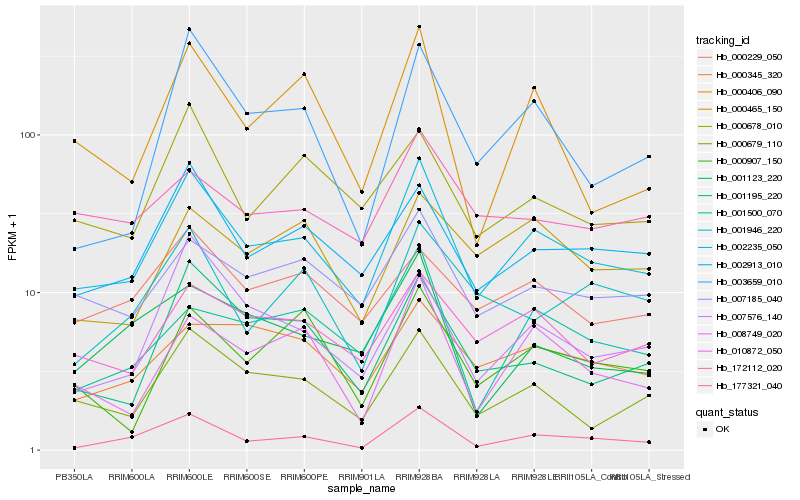
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_050 |
0.0 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002913_010 |
0.1124296706 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000679_110 |
0.1156795642 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 4 |
Hb_177321_040 |
0.1231430092 |
- |
- |
hypothetical protein JCGZ_06328 [Jatropha curcas] |
| 5 |
Hb_010872_050 |
0.1284843323 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 6 |
Hb_007185_040 |
0.1295426656 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 7 |
Hb_000678_010 |
0.1391326147 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_008749_020 |
0.1397294275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007576_140 |
0.1416788671 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 10 |
Hb_001195_220 |
0.1425206556 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_003659_010 |
0.1454834615 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 12 |
Hb_000229_050 |
0.1470532182 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 13 |
Hb_000406_090 |
0.1498444413 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_001123_220 |
0.1548280381 |
- |
- |
- |
| 15 |
Hb_000345_320 |
0.156650242 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000907_150 |
0.1571071963 |
- |
- |
hypothetical protein L484_004295 [Morus notabilis] |
| 17 |
Hb_172112_020 |
0.1576982732 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 18 |
Hb_001500_070 |
0.1580170853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000465_150 |
0.1601610633 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 20 |
Hb_001946_220 |
0.1605448242 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |