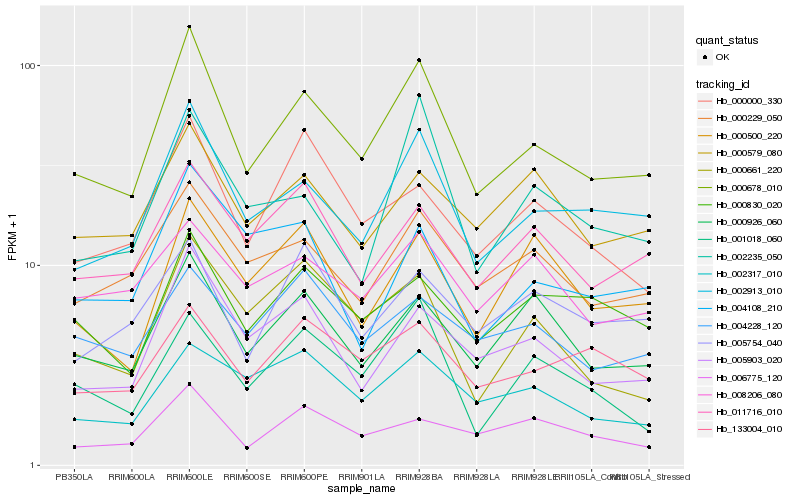
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000678_010 |
0.0 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002913_010 |
0.0820677317 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005903_020 |
0.1149890915 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 4 |
Hb_000229_050 |
0.1191483468 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 5 |
Hb_000926_060 |
0.1192843785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011716_010 |
0.1282279951 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 7 |
Hb_000830_020 |
0.1323638344 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 8 |
Hb_000661_220 |
0.1363997886 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 9 |
Hb_000579_080 |
0.137073314 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 10 |
Hb_006775_120 |
0.1384816817 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 11 |
Hb_133004_010 |
0.1389915418 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 12 |
Hb_002235_050 |
0.1391326147 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000500_220 |
0.1406181813 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 14 |
Hb_000000_330 |
0.1406550537 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 15 |
Hb_001018_060 |
0.1416730623 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 16 |
Hb_002317_010 |
0.1422395477 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_004108_210 |
0.1453123366 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 18 |
Hb_004228_120 |
0.1459914627 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 19 |
Hb_008206_080 |
0.1472150598 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 20 |
Hb_005754_040 |
0.1480693861 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |