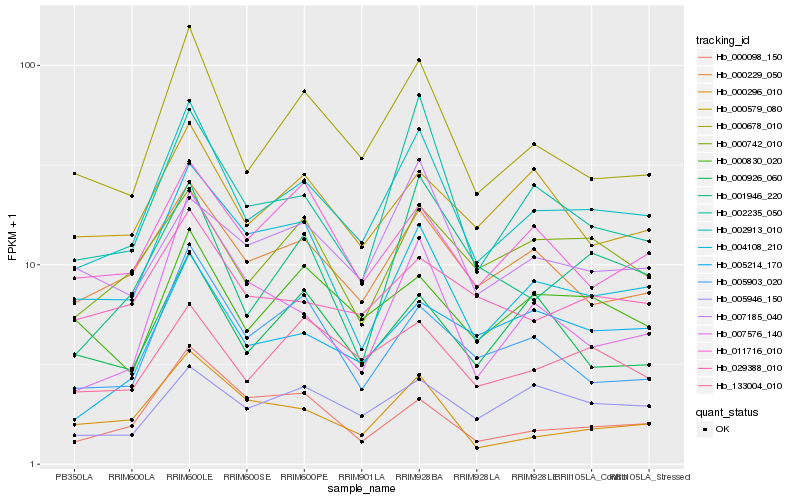
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002913_010 |
0.0 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000678_010 |
0.0820677317 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000229_050 |
0.10822287 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 4 |
Hb_002235_050 |
0.1124296706 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000296_010 |
0.1285700819 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 6 |
Hb_000742_010 |
0.1310172501 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_133004_010 |
0.1312672094 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 8 |
Hb_029388_010 |
0.1325653339 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 9 |
Hb_004108_210 |
0.1326064594 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 10 |
Hb_005903_020 |
0.1342848047 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 11 |
Hb_011716_010 |
0.1349879727 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 12 |
Hb_000830_020 |
0.1359420443 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 13 |
Hb_007185_040 |
0.1367285484 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 14 |
Hb_000579_080 |
0.1374513598 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 15 |
Hb_005214_170 |
0.1375641833 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 16 |
Hb_001946_220 |
0.1375806166 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 17 |
Hb_007576_140 |
0.1383128444 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 18 |
Hb_005946_150 |
0.1389794552 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 19 |
Hb_000098_150 |
0.139452147 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 20 |
Hb_000926_060 |
0.1416294813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |