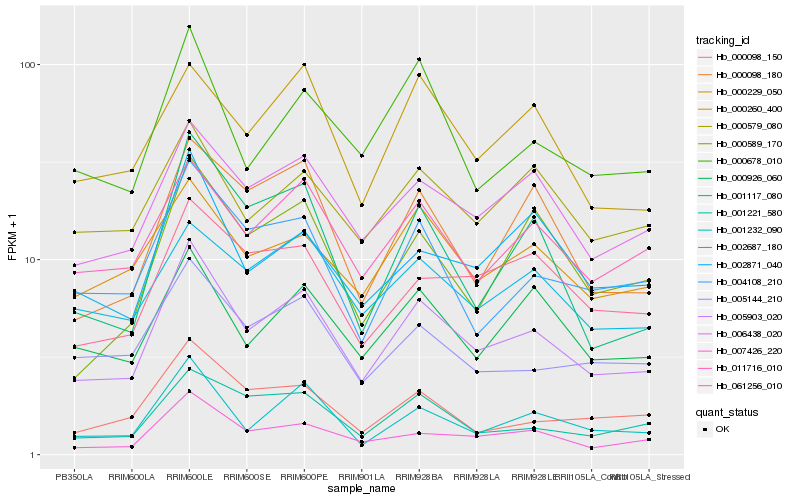
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005903_020 |
0.0 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 2 |
Hb_006438_020 |
0.0992904208 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 3 |
Hb_007426_220 |
0.0998352878 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_001232_090 |
0.1052561059 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 5 |
Hb_011716_010 |
0.1073718111 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 6 |
Hb_004108_210 |
0.1113621868 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 7 |
Hb_000926_060 |
0.1123416822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000678_010 |
0.1149890915 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_001221_580 |
0.1151819362 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 10 |
Hb_005144_210 |
0.1166186814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000229_050 |
0.1172939098 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 12 |
Hb_061256_010 |
0.1193445182 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000098_180 |
0.1194176997 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000098_150 |
0.1207875569 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 15 |
Hb_002871_040 |
0.121184562 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 16 |
Hb_001117_080 |
0.1219054128 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_400 |
0.1226614457 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 18 |
Hb_002687_180 |
0.1228179606 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 19 |
Hb_000589_170 |
0.1236840349 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000579_080 |
0.1244505569 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |