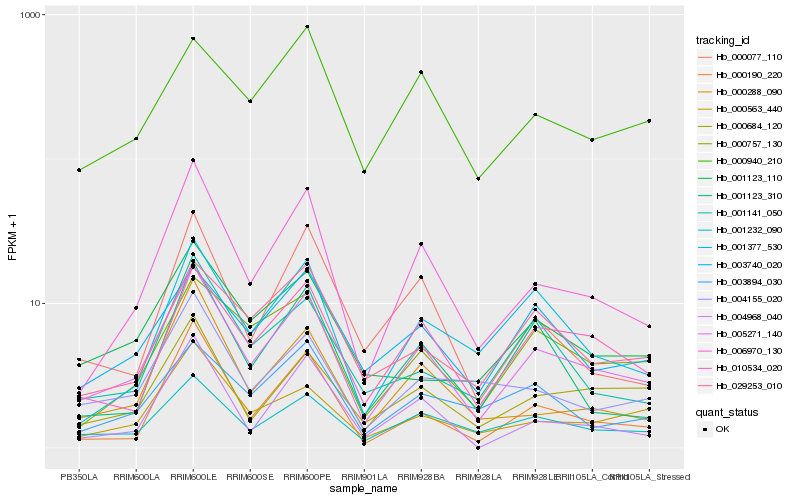
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_140 |
0.0 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 2 |
Hb_006970_130 |
0.101009666 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 3 |
Hb_001232_090 |
0.110642799 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 4 |
Hb_010534_020 |
0.1197580868 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000684_120 |
0.131932665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000757_130 |
0.1371757346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000563_440 |
0.1408478404 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003740_020 |
0.1451649678 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_000190_220 |
0.1460878761 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 10 |
Hb_029253_010 |
0.1462926925 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 11 |
Hb_001377_530 |
0.148786252 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_001141_050 |
0.1529840375 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003894_030 |
0.1546381556 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 14 |
Hb_001123_110 |
0.1547762189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004155_020 |
0.155248016 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_000077_110 |
0.1566275334 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 17 |
Hb_000288_090 |
0.1568960032 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001123_310 |
0.1570559663 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 19 |
Hb_000940_210 |
0.1580808629 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 20 |
Hb_004968_040 |
0.1609909916 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |