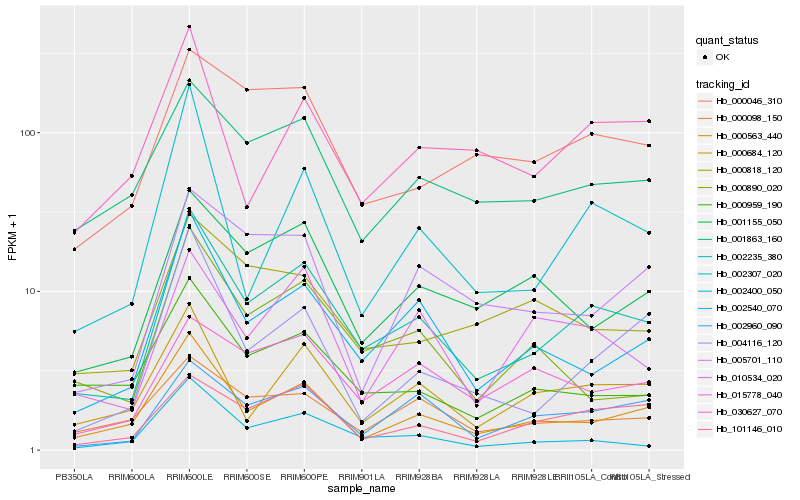
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_380 |
0.0 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 2 |
Hb_000563_440 |
0.1321746433 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002540_070 |
0.136927572 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 4 |
Hb_000684_120 |
0.1551670646 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002960_090 |
0.1562312789 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001863_160 |
0.1587001926 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 7 |
Hb_001155_050 |
0.1616120742 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 8 |
Hb_101146_010 |
0.1624268168 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 9 |
Hb_002400_050 |
0.1661179975 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000046_310 |
0.1685341291 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 11 |
Hb_000098_150 |
0.1687096589 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 12 |
Hb_000959_190 |
0.1689096095 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 13 |
Hb_010534_020 |
0.1717333021 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002307_020 |
0.1726789315 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 15 |
Hb_030627_070 |
0.1772203476 |
- |
- |
histone H4 [Zea mays] |
| 16 |
Hb_000818_120 |
0.1778771917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000890_020 |
0.1781975788 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_005701_110 |
0.1799301941 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 19 |
Hb_004116_120 |
0.1800352943 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 20 |
Hb_015778_040 |
0.1806821632 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |