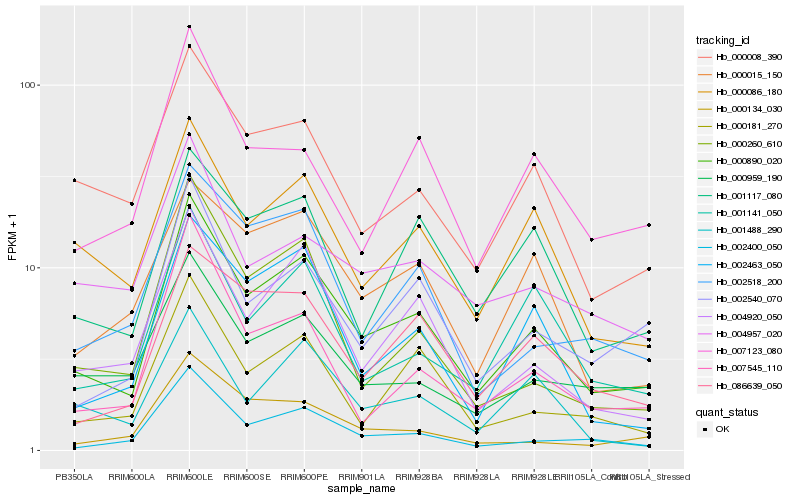
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_020 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_004920_050 |
0.1257861888 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 3 |
Hb_002400_050 |
0.1266372035 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002540_070 |
0.1328677815 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 5 |
Hb_000086_180 |
0.1337074619 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 6 |
Hb_000008_390 |
0.1365970556 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 7 |
Hb_000181_270 |
0.1381134204 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002518_200 |
0.1408492361 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 9 |
Hb_002463_050 |
0.1413220487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001141_050 |
0.1417144976 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_086639_050 |
0.1438993199 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000959_190 |
0.1461729947 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 13 |
Hb_001117_080 |
0.1478500865 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000015_150 |
0.1493458769 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 15 |
Hb_000260_610 |
0.1504145896 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 16 |
Hb_007545_110 |
0.151222633 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_001488_290 |
0.1512251828 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000134_030 |
0.1518700235 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 19 |
Hb_007123_080 |
0.1536560561 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 20 |
Hb_004957_020 |
0.1573486407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |