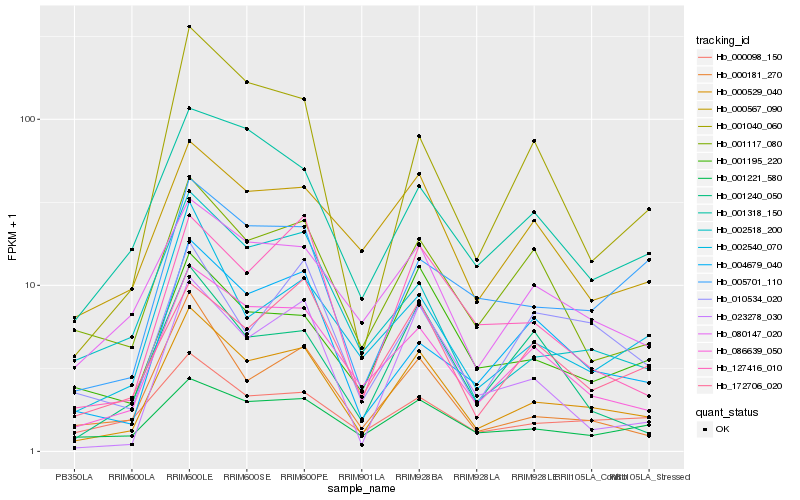
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 2 |
Hb_086639_050 |
0.1166518846 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000181_270 |
0.1334058477 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_080147_020 |
0.137908756 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 5 |
Hb_001117_080 |
0.1473150753 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002518_200 |
0.1496915004 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 7 |
Hb_000567_090 |
0.150507754 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002540_070 |
0.1509756451 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 9 |
Hb_172706_020 |
0.1512543452 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 10 |
Hb_001195_220 |
0.151380937 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_001221_580 |
0.1517156602 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 12 |
Hb_127416_010 |
0.1531921751 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_004679_040 |
0.1579986491 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 14 |
Hb_001318_150 |
0.1588343266 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 15 |
Hb_001240_050 |
0.1602413894 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 16 |
Hb_023278_030 |
0.1608993455 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000098_150 |
0.1610521694 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 18 |
Hb_005701_110 |
0.1610819435 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 19 |
Hb_001040_060 |
0.164200042 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 20 |
Hb_010534_020 |
0.1647975862 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |