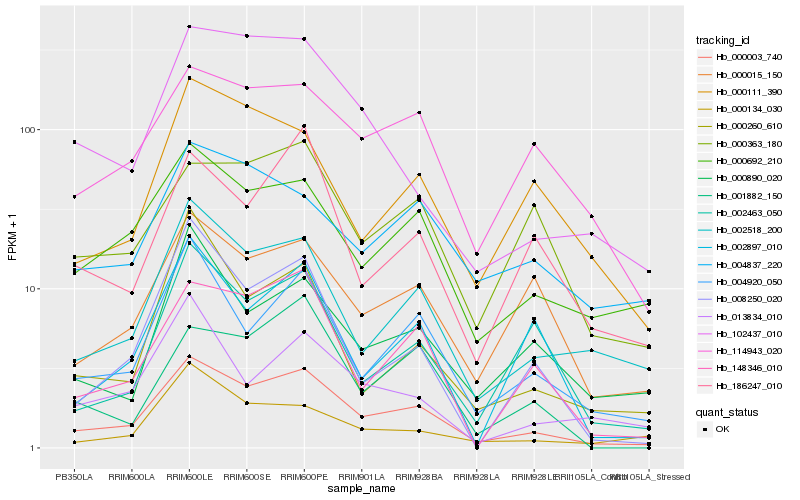
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_740 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 2 |
Hb_002518_200 |
0.1328668905 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 3 |
Hb_000692_210 |
0.1358530957 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_004920_050 |
0.1393294345 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 5 |
Hb_148346_010 |
0.1409688415 |
- |
- |
- |
| 6 |
Hb_114943_020 |
0.1507937937 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 7 |
Hb_000015_150 |
0.1520613998 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 8 |
Hb_002463_050 |
0.1655521077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008250_020 |
0.1700062242 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000890_020 |
0.1707062211 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000111_390 |
0.1712311106 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 12 |
Hb_102437_010 |
0.1727490127 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_000363_180 |
0.1736167693 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 14 |
Hb_186247_010 |
0.1736877972 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 15 |
Hb_002897_010 |
0.1741759548 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 16 |
Hb_001882_150 |
0.1742855288 |
transcription factor |
TF Family: mTERF |
- |
| 17 |
Hb_004837_220 |
0.1800554241 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 18 |
Hb_000134_030 |
0.181183829 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 19 |
Hb_013834_010 |
0.1811925087 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 20 |
Hb_000260_610 |
0.1851867557 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |