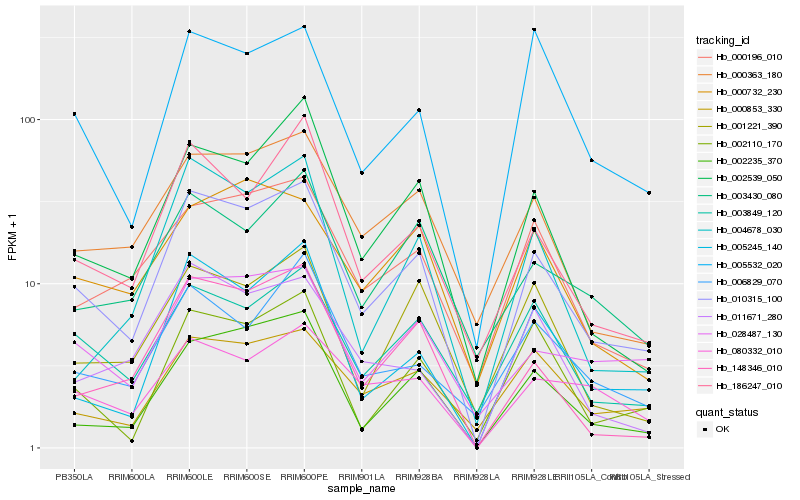
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010315_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 2 |
Hb_000363_180 |
0.1255745009 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 3 |
Hb_002235_370 |
0.1267053001 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 4 |
Hb_002539_050 |
0.1272484281 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 5 |
Hb_003849_120 |
0.1319083552 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 6 |
Hb_186247_010 |
0.13461037 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 7 |
Hb_000196_010 |
0.1362519885 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 8 |
Hb_028487_130 |
0.1367524549 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 9 |
Hb_005245_140 |
0.137919693 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 10 |
Hb_148346_010 |
0.1389664181 |
- |
- |
- |
| 11 |
Hb_000853_330 |
0.1449155787 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_080332_010 |
0.145593859 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 13 |
Hb_003430_080 |
0.1483464375 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002110_170 |
0.1483503236 |
- |
- |
hypothetical protein CICLE_v10002406mg [Citrus clementina] |
| 15 |
Hb_005532_020 |
0.1527202926 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 16 |
Hb_000732_230 |
0.1529592742 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011671_280 |
0.1534075899 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 18 |
Hb_004678_030 |
0.1540021046 |
- |
- |
fructokinase [Manihot esculenta] |
| 19 |
Hb_001221_390 |
0.1551696554 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 20 |
Hb_006829_070 |
0.1555320986 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |