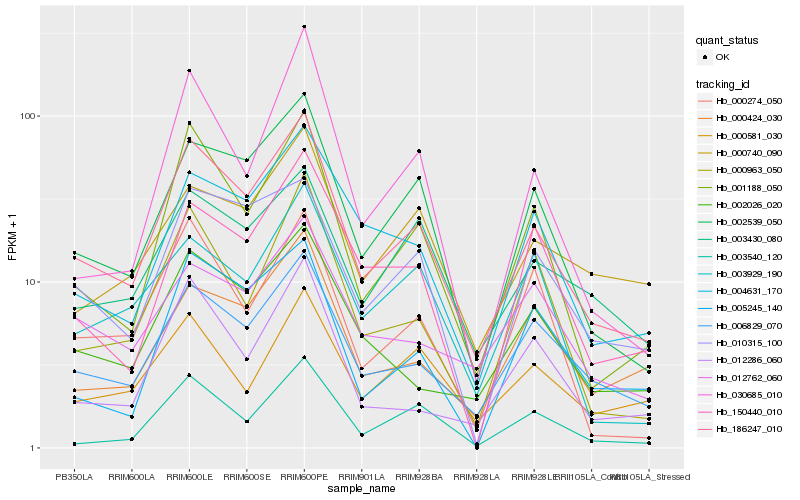
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186247_010 |
0.0 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 2 |
Hb_002539_050 |
0.1032117499 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 3 |
Hb_001188_050 |
0.1058888632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006829_070 |
0.1061253126 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 5 |
Hb_010315_100 |
0.13461037 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 6 |
Hb_004631_170 |
0.1347819284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002026_020 |
0.1357663117 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 8 |
Hb_003430_080 |
0.1367894724 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_003540_120 |
0.1456366357 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_000424_030 |
0.1489087117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000963_050 |
0.1490618204 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 12 |
Hb_012286_060 |
0.1498539549 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_012762_060 |
0.1508846021 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_000274_050 |
0.1518463312 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_005245_140 |
0.1527326094 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 16 |
Hb_150440_010 |
0.1531653134 |
- |
- |
- |
| 17 |
Hb_000740_090 |
0.1537364164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_030685_010 |
0.1554645533 |
- |
- |
- |
| 19 |
Hb_000581_030 |
0.1561854625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003929_190 |
0.1573816765 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |