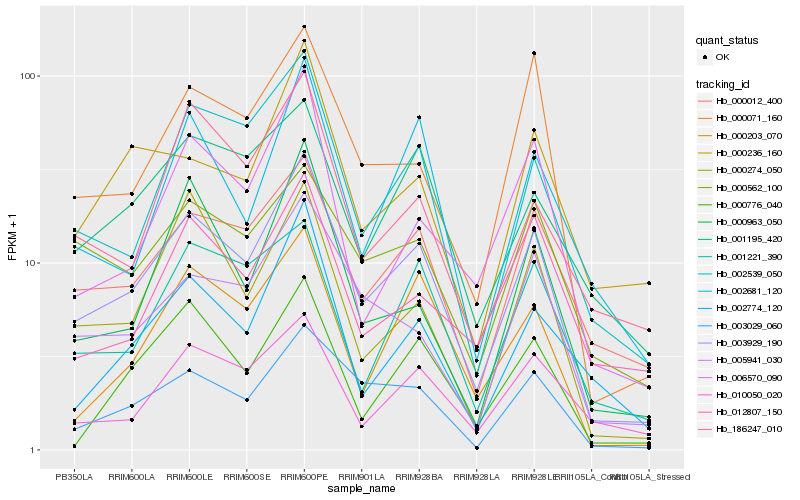
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_190 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 2 |
Hb_000012_400 |
0.116184396 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 3 |
Hb_002539_050 |
0.1212443978 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 4 |
Hb_003029_060 |
0.1214535783 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005941_030 |
0.1383090515 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000071_160 |
0.1405422368 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_000203_070 |
0.1434937419 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 8 |
Hb_001195_420 |
0.1465511713 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 9 |
Hb_010050_020 |
0.1473669549 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 10 |
Hb_006570_090 |
0.1486057997 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 11 |
Hb_002681_120 |
0.1494187646 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 12 |
Hb_000274_050 |
0.1496980699 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 13 |
Hb_002774_120 |
0.1515923062 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 14 |
Hb_000562_100 |
0.1538065961 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 15 |
Hb_001221_390 |
0.1545584605 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 16 |
Hb_000776_040 |
0.1560164304 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000236_160 |
0.15688635 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 18 |
Hb_012807_150 |
0.1572403774 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 19 |
Hb_186247_010 |
0.1573816765 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 20 |
Hb_000963_050 |
0.1580987382 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |