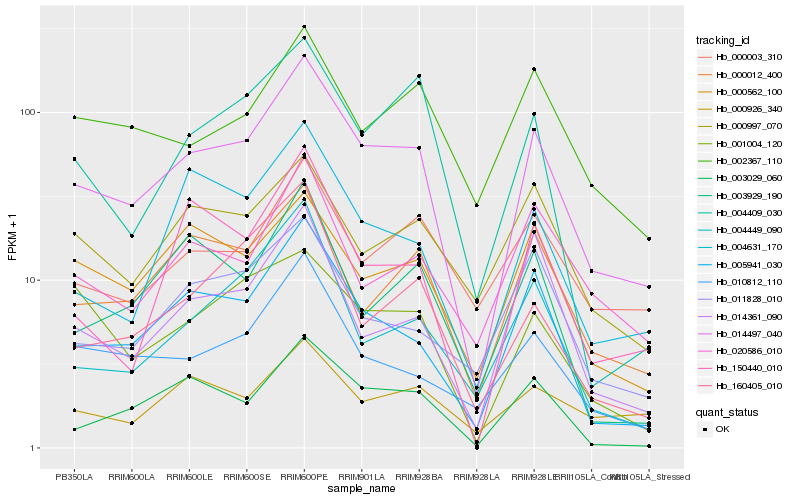
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_040 |
0.0 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_005941_030 |
0.1138434631 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000012_400 |
0.1345701613 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 4 |
Hb_003029_060 |
0.1416441411 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010812_110 |
0.1489527223 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 6 |
Hb_000562_100 |
0.1515938889 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 7 |
Hb_004631_170 |
0.1519244984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_014361_090 |
0.1547396381 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 9 |
Hb_020586_010 |
0.1550396955 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_011828_010 |
0.1589803267 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 11 |
Hb_000003_310 |
0.1591220678 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 12 |
Hb_002367_110 |
0.1603642507 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 13 |
Hb_004409_030 |
0.1604712 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 14 |
Hb_000997_070 |
0.1611927143 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 15 |
Hb_003929_190 |
0.1619850759 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 16 |
Hb_150440_010 |
0.1621702222 |
- |
- |
- |
| 17 |
Hb_004449_090 |
0.164232746 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 18 |
Hb_001004_120 |
0.165104315 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000926_340 |
0.1675603402 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_160405_010 |
0.1687685177 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |