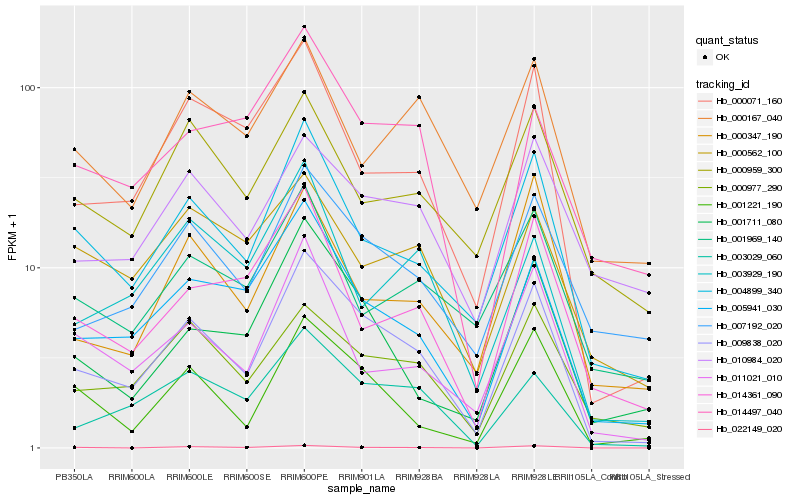
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_020 |
0.0 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 2 |
Hb_004899_340 |
0.1304492328 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 3 |
Hb_005941_030 |
0.1308459557 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_003029_060 |
0.1349470196 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001221_190 |
0.1467618964 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 6 |
Hb_000071_160 |
0.1487554749 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_000977_290 |
0.1547795929 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_007192_020 |
0.1551675832 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 9 |
Hb_001711_080 |
0.1584101148 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 10 |
Hb_011021_010 |
0.1626361828 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_014497_040 |
0.1764075158 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_014361_090 |
0.1775162384 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 13 |
Hb_000347_190 |
0.1789294953 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 14 |
Hb_010984_020 |
0.1843329302 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 15 |
Hb_000959_300 |
0.1865480414 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 16 |
Hb_003929_190 |
0.1880580853 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 17 |
Hb_000167_040 |
0.1925004561 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 18 |
Hb_000562_100 |
0.1933757734 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 19 |
Hb_001969_140 |
0.1934071406 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 20 |
Hb_022149_020 |
0.1935146016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |