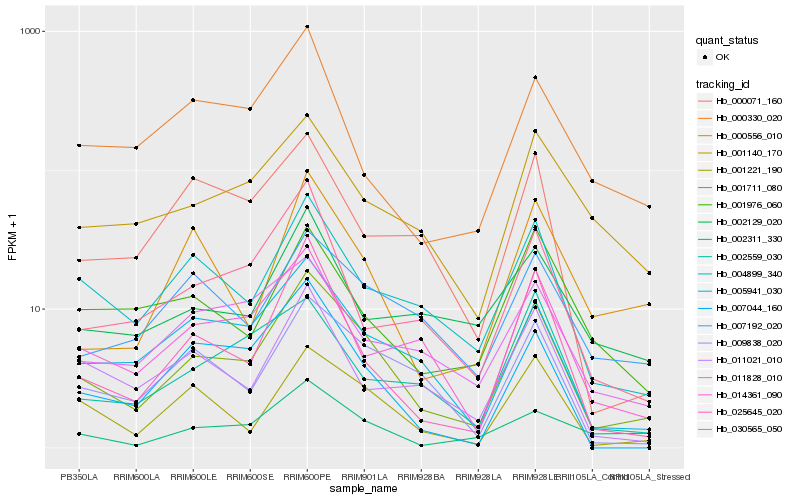
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001711_080 |
0.0 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 2 |
Hb_005941_030 |
0.1527869178 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_004899_340 |
0.1531923122 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 4 |
Hb_001221_190 |
0.1567797191 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 5 |
Hb_009838_020 |
0.1584101148 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 6 |
Hb_025645_020 |
0.163464747 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 7 |
Hb_014361_090 |
0.17205335 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 8 |
Hb_007044_160 |
0.1768570583 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 9 |
Hb_007192_020 |
0.1808104171 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 10 |
Hb_030565_050 |
0.1810448412 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 11 |
Hb_011828_010 |
0.1835138469 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 12 |
Hb_001140_170 |
0.1835698414 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 13 |
Hb_002311_330 |
0.1839959662 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_011021_010 |
0.1883665312 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000556_010 |
0.1895760207 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000071_160 |
0.1944915474 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 17 |
Hb_002559_030 |
0.1974871264 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 18 |
Hb_000330_020 |
0.199766197 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 19 |
Hb_002129_020 |
0.1997927224 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 20 |
Hb_001976_060 |
0.2021939143 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |