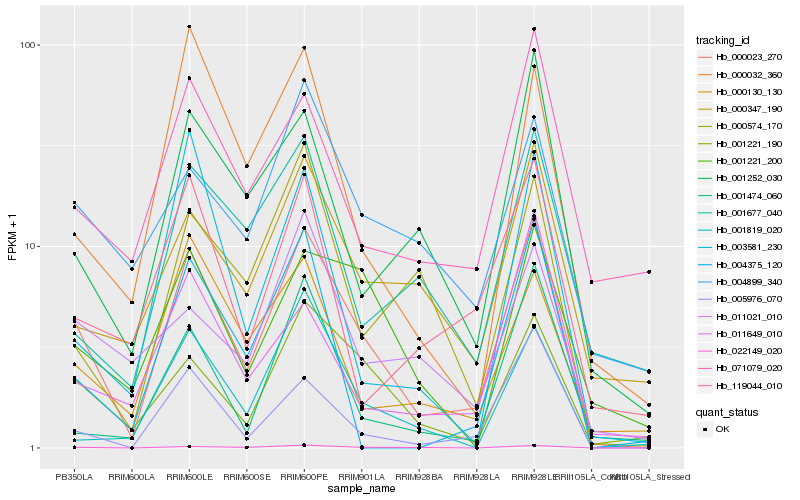
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_022149_020 |
0.1613837243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001221_190 |
0.1764012957 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 4 |
Hb_005976_070 |
0.1828237969 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 5 |
Hb_001252_030 |
0.2015751597 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 6 |
Hb_000347_190 |
0.2041877178 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 7 |
Hb_071079_020 |
0.2111230162 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 8 |
Hb_000130_130 |
0.2125294655 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 9 |
Hb_001221_200 |
0.2134890946 |
- |
- |
- |
| 10 |
Hb_119044_010 |
0.2151966213 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 11 |
Hb_000032_360 |
0.2152273481 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004899_340 |
0.2156254539 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_001474_060 |
0.2200798947 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 14 |
Hb_001819_020 |
0.2224363795 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004375_120 |
0.2227971144 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 3-like [Jatropha curcas] |
| 16 |
Hb_001677_040 |
0.2236399693 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011649_010 |
0.2237070674 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 18 |
Hb_000574_170 |
0.224392237 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 19 |
Hb_011021_010 |
0.2259297063 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_003581_230 |
0.2259588789 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |