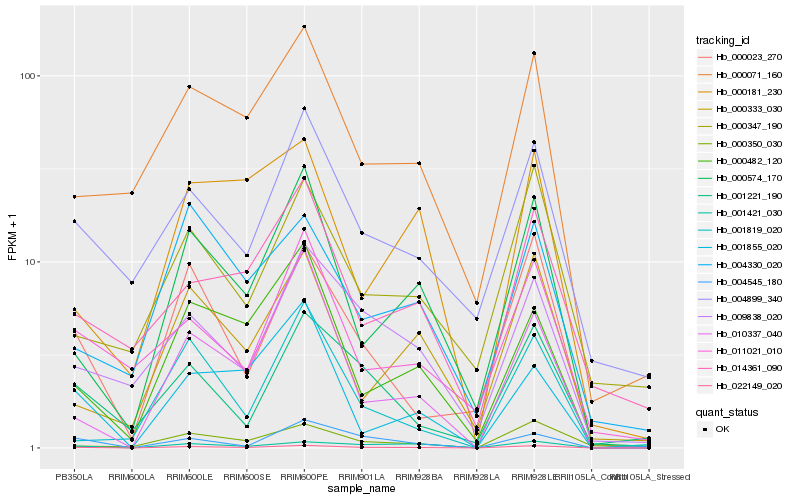
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022149_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000574_170 |
0.152770057 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 3 |
Hb_000023_270 |
0.1613837243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001421_030 |
0.1695785955 |
- |
- |
- |
| 5 |
Hb_010337_040 |
0.1870428493 |
- |
- |
- |
| 6 |
Hb_000333_030 |
0.1874641913 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000482_120 |
0.1876714183 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 8 |
Hb_001221_190 |
0.1895750231 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 9 |
Hb_000071_160 |
0.1900419466 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 10 |
Hb_009838_020 |
0.1935146016 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 11 |
Hb_000350_030 |
0.2015207901 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 12 |
Hb_004899_340 |
0.2025770091 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_000347_190 |
0.2038656735 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 14 |
Hb_004545_180 |
0.2068733745 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_011021_010 |
0.2094951676 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_004330_020 |
0.2096522498 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 17 |
Hb_001819_020 |
0.2114088143 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000181_230 |
0.2142827585 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_014361_090 |
0.2146513438 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 20 |
Hb_001855_020 |
0.2162060692 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |