| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
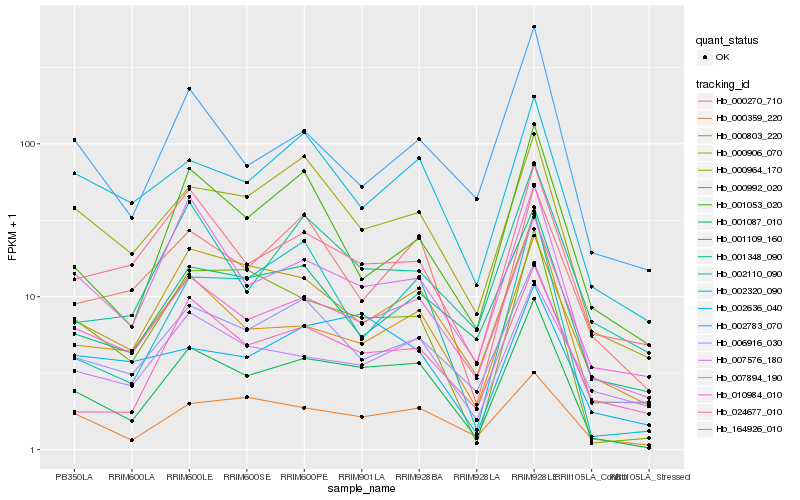
Hb_001087_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630042 [Jatropha curcas] |
| 2 |
Hb_000906_070 |
0.1358014319 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000992_020 |
0.1427224358 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 4 |
Hb_010984_010 |
0.1441832332 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000803_220 |
0.1504020711 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007894_190 |
0.1548966059 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 7 |
Hb_000359_220 |
0.1552062463 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000964_170 |
0.155901889 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 9 |
Hb_024677_010 |
0.1582743127 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 10 |
Hb_002320_090 |
0.1590961974 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 11 |
Hb_006916_030 |
0.1603773932 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 12 |
Hb_000270_710 |
0.1647591416 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001109_160 |
0.1652562172 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002110_090 |
0.1669215634 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 15 |
Hb_007576_180 |
0.1674807201 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 16 |
Hb_164926_010 |
0.167991545 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 17 |
Hb_002783_070 |
0.1721910475 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 18 |
Hb_001053_020 |
0.1741436813 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 19 |
Hb_001348_090 |
0.1766642583 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002636_040 |
0.1788670485 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |