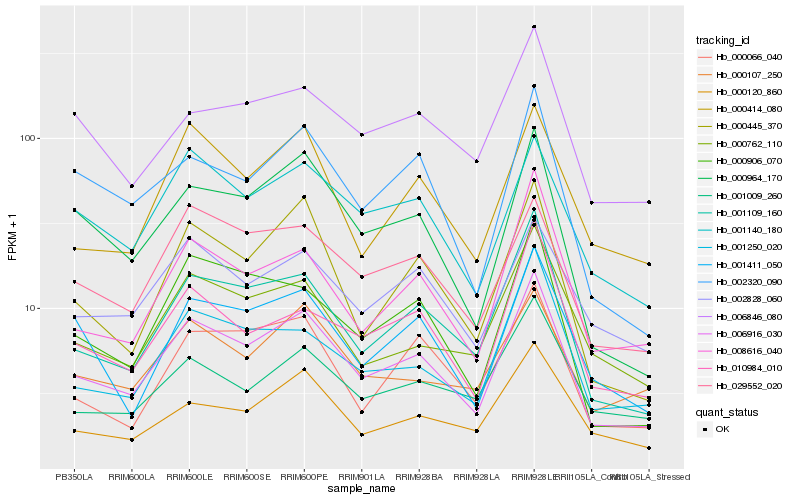
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 2 |
Hb_001109_160 |
0.0787900924 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000964_170 |
0.0926268715 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_001140_180 |
0.0989092303 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 5 |
Hb_000445_370 |
0.1013642166 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 6 |
Hb_008616_040 |
0.10222164 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 7 |
Hb_029552_020 |
0.1027888355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000762_110 |
0.1032780541 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002320_090 |
0.104485026 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 10 |
Hb_010984_010 |
0.1062648862 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000906_070 |
0.1070887513 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001250_020 |
0.1075321775 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001411_050 |
0.1080323054 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 14 |
Hb_000414_080 |
0.1080615418 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000066_040 |
0.1100671552 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 16 |
Hb_000120_860 |
0.1110237092 |
- |
- |
nucellin, putative [Ricinus communis] |
| 17 |
Hb_002828_060 |
0.1121093121 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000107_250 |
0.1135209171 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 19 |
Hb_006846_080 |
0.11401864 |
- |
- |
calnexin, putative [Ricinus communis] |
| 20 |
Hb_001009_260 |
0.1166177821 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |