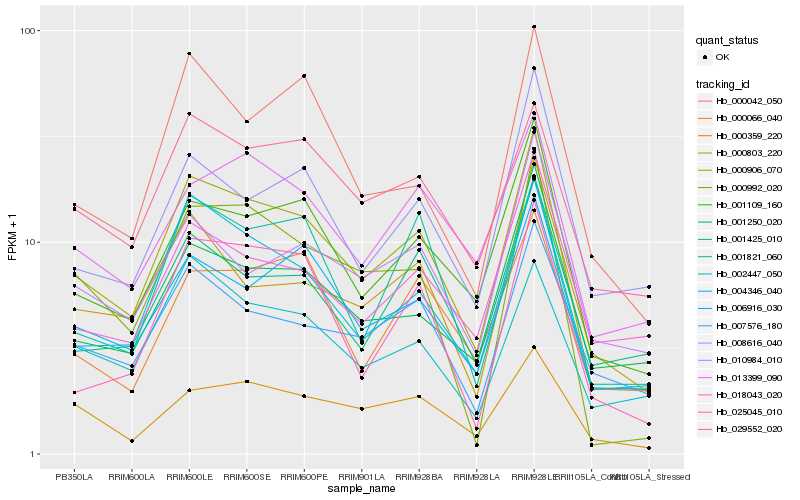
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000906_070 |
0.0 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004346_040 |
0.0947410544 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 3 |
Hb_001109_160 |
0.0976027131 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_006916_030 |
0.1070887513 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 5 |
Hb_000359_220 |
0.116032722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 6 |
Hb_029552_020 |
0.1208088398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_013399_090 |
0.122110955 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_007576_180 |
0.1226064641 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 9 |
Hb_001250_020 |
0.1237769342 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000992_020 |
0.1242520997 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 11 |
Hb_002447_050 |
0.1243186783 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000803_220 |
0.1263911862 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000042_050 |
0.1274743841 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001821_060 |
0.1280743103 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 15 |
Hb_025045_010 |
0.1303279566 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_001425_010 |
0.1313895225 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_000066_040 |
0.1324833973 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 18 |
Hb_018043_020 |
0.1339122914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_008616_040 |
0.1344705997 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010984_010 |
0.1347465547 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |