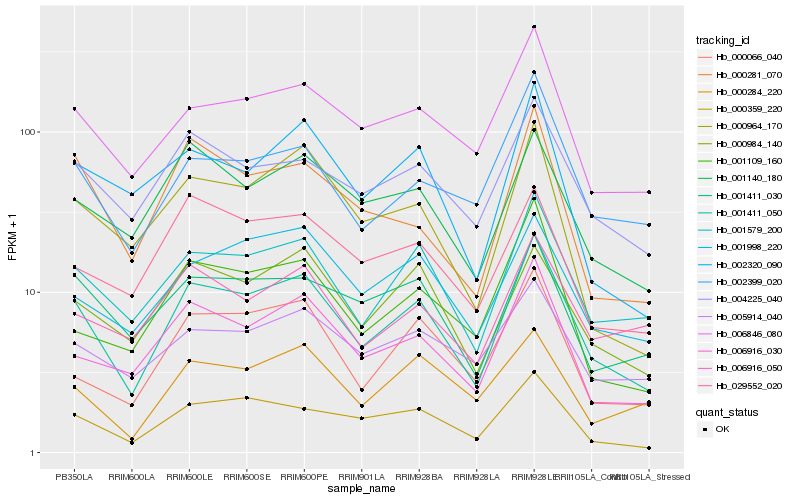
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001411_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 2 |
Hb_001579_200 |
0.0923230254 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 3 |
Hb_000359_220 |
0.107245811 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006916_030 |
0.1080323054 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 5 |
Hb_006846_080 |
0.1137941126 |
- |
- |
calnexin, putative [Ricinus communis] |
| 6 |
Hb_004225_040 |
0.115375386 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 7 |
Hb_001140_180 |
0.122416953 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 8 |
Hb_000984_140 |
0.1226968411 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 9 |
Hb_000964_170 |
0.1228893853 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 10 |
Hb_000066_040 |
0.1232862842 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 11 |
Hb_001411_030 |
0.1239114323 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 12 |
Hb_001998_220 |
0.1263616665 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002399_020 |
0.1270488035 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 14 |
Hb_002320_090 |
0.1294737001 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 15 |
Hb_001109_160 |
0.1321439627 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_005914_040 |
0.1336960699 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 17 |
Hb_029552_020 |
0.1342454469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000281_070 |
0.1345846006 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 19 |
Hb_000284_220 |
0.1360765738 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 20 |
Hb_006916_050 |
0.1365909756 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |