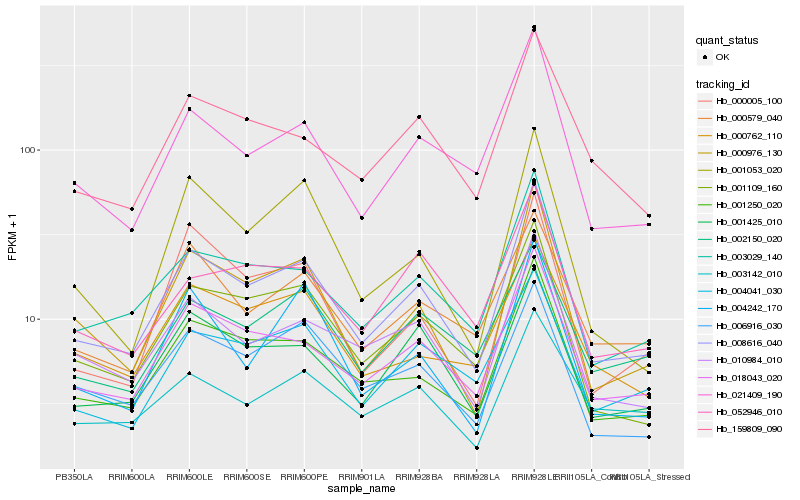
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008616_040 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 2 |
Hb_018043_020 |
0.0713503786 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001250_020 |
0.0715318365 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003029_140 |
0.0758186927 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000976_130 |
0.0791421211 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 6 |
Hb_021409_190 |
0.0844581437 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 7 |
Hb_010984_010 |
0.0855662724 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001109_160 |
0.0860467379 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001425_010 |
0.0881948463 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_006916_030 |
0.10222164 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 11 |
Hb_159809_090 |
0.1041821875 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 12 |
Hb_004041_030 |
0.1051405702 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000579_040 |
0.1107857368 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 14 |
Hb_052946_010 |
0.1137432313 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 15 |
Hb_000762_110 |
0.1141655183 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004242_170 |
0.1150278714 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 17 |
Hb_002150_020 |
0.1154547835 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_001053_020 |
0.1157831958 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 19 |
Hb_003142_010 |
0.1163553801 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 20 |
Hb_000005_100 |
0.1170665847 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |